Abstract
Embryonic stem (ES) cell self-renewal is regulated by transcription factors, including Oct4, Sox2, and Nanog. A number of additional transcriptional regulators of ES cell self-renewal have recently been identified, including the orphan nuclear receptor estrogen-related receptor beta (Esrrb). However, the mode of action of Esrrb in ES cells is unknown. Here, using an Oct4 affinity screen, we identify Esrrb as an Oct4 partner protein. Esrrb can interact with Oct4 independently of DNA. Esrrb is recruited near the Oct-Sox element in the Nanog proximal promoter, where it positively regulates Nanog expression. Esrrb recruitment to the Nanog promoter requires both the presence of Oct4 and a degenerate estrogen-related receptor DNA element. Consistent with its role in Nanog regulation, expression of the Esrrb protein within the Oct4-positive ES cell population is mosaic and correlates with the mosaic expression of the Nanog protein. Together with previous reports that Nanog may regulate Esrrb gene expression, our results suggest that Esrrb and Nanog act as part of a feedback regulatory circuit that modulates the fluctuating self-renewal capacity of ES cell populations.
The self-renewal of mouse ES cells is regulated by a network of transcription factors that includes Oct4, Nanog, and Sox2 (22). The expression level of Oct4 protein needs to be kept within a tight range in order to maintain ES cell self-renewal (23). Decreasing Oct4 levels below 50% induces differentiation into the trophectoderm, whereas a twofold increase causes differentiation into cells expressing markers of the endoderm and mesoderm (23). In contrast, overexpression of Nanog allows mouse embryonic stem (ES) cells to remain undifferentiated in the absence of the otherwise requisite stimulation by leukemia inhibitory factor and bone morphogenetic protein (5, 19, 34). Oct4 is thought to act together with Sox2 by binding to adjacent cognate DNA sequences in many genes (1), including Nanog (14, 26). Genome-wide chromatin immunoprecipitation (ChIP) studies have suggested that composite Oct-Sox motifs regulate the expression of many genes in mouse and human ES cells (2, 16). Recent evidence has shown that the critical role of Sox2 in maintaining ES cell self-renewal is regulating Oct4 expression, suggesting that the secondary role of gene regulation via Oct-Sox motifs is performed redundantly with Sox4, Sox11, and Sox15 (18).
Recent reports have expanded the list of factors that contribute to ES cell self-renewal. Wang et al. (30) reported a proteomic analysis of interactors of Oct4 and Nanog and suggested that some Nanog interactors may assist in Nanog-mediated gene regulation. A separate study using an RNA interference (RNAi) screen found that depletion of estrogen-related receptor beta (Esrrb), Tbx3, or Tcl1 resulted in ES cell differentiation but that this differentiation could be attenuated by overexpression of Nanog (13). However, it is unclear how any of these novel regulatory factors mediate their function. Here we use an unbiased analysis of Oct4 binding proteins to identify Esrrb as an Oct4-interacting partner protein. We show that Esrrb is recruited to the Oct4 responsive element within the proximal Nanog promoter where it is responsible for mediating the positive regulatory effect of Oct4.
MATERIALS AND METHODS
Plasmids and cell culture.
RNAi constructs pSuper-Esrrb-sh1 and pSuper-Esrrb-sh2 were constructed by cloning Esrrb RNAi1 and RNAi2 (16) into pSuper-puro (Oligoengine). pSuper-control contains an oligonucleotide without complementarity to any known mammalian sequence (Dharmacon). Mouse ES cell lines 46C (35) and ZHBTc4 (23) and derivatives of ZHBTc4 were grown on gelatin-coated dishes without feeders on Glasgow minimal essential medium supplemented with leukemia inhibitory factor, 15% fetal bovine serum, 0.25% sodium bicarbonate, 1 mM glutamine, 1 mM sodium pyruvate, nonessential amino acids, 50 μM beta-mercaptoethanol, and penicillin-streptomycin.
Cells from the c6 cell line, called F-Oct4 ES cells from here onwards, were created by electroporating ZHBTc4 cells with linearized pPyCAG (FLAG)3 Oct4IP, a plasmid in which the Oct4 open reading frame was placed between the N-terminal triple FLAG tag and the internal ribosome entry site (IRES)-puromycin resistance cassette of pPyCAG (FLAG)3IP (20). Electroporated cells were plated in ES cell medium, and after 24 h, 1 μg/ml puromycin and 1 μg/ml doxycycline were added. After 12 days of selection, puromycin-resistant colonies were picked and tested for FLAG-Oct4 expression by anti-FLAG Western blot analysis. TNG cells have been described previously (6) Puromycin-sensitive TNG-PS cells were derived from TNG cells by excision of the frt-IRES-pac-frt cassette by transient expression of FLPe. Bright field pictures of ES cell cultures were taken using the IX70 inverted microscope (Olympus).
Oct4 purification and mass spectrometry.
F-Oct4 ES cells and control cells (ZHBTc4) were expanded to 50 14-cm dishes, plates were washed once with phosphate-buffered saline, cells were scraped off, and nuclear extracts were prepared (8) and dialyzed to 100 mM KCl (8). A total of 100 μl of anti-FLAG M2 agarose beads (Sigma), equilibrated in buffer C-100 (20 mM HEPES, pH 7.6, 10% glycerol, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1× complete EDTA-free protease inhibitor; Roche), was added to 10 ml of nuclear extract, incubated for 3 h at 4°C, transferred into an Eppendorf tube, and washed five times with 1 ml of C-100 buffer plus 0.02% NP-40 (C-100*) and eluted four times with C-100* containing 0.2 mg/ml FLAG tripeptide (Sigma) for 15 min at 4°C. Fractions were loaded onto a 10% sodium dodecyl sulfate (SDS)-phosphonoacetic acid (PAA) gel and silver stained. Elutions 1 and 2, containing the majority of FLAG-Oct4 in purification from the F-Oct4 extract, were concentrated by SpeedVac condensation, loaded onto a 10% SDS-PAA gel, and stained with colloidal Coomassie blue. Gel lanes were cut and subjected to in-gel digestion with trypsin (Promega), essentially as described previously (31). Nanoflow liquid chromatography-tandem mass spectrometry was performed on a 1100 series capillary liquid chromatography system (Agilent Technologies) coupled to an LTQ-Orbitrap mass spectrometer (Thermo), as described previously (27). Database searches to assign proteins to the found peptide fragmentation spectra were performed using Mascot, as described previously (27).
IP.
For immunoprecipitation (IP), 2.5 μg of Oct4 antibody (N19; Santa Cruz Biotechnology) or Esrrb antibody (R&D Systems) was added to 200 μl of 46C ES cell nuclear extract and incubated under rotation for 2 h at 4°C. A total of 1 U Benzonase (Novagen) or 25 μg/ml ethidium bromide was added where indicated. The antibody-extract mixture was added to 20 μl of protein G-Sepharose beads (Amersham) blocked with 1% fish skin gelatin (Sigma) and 0.2 mg/ml chicken egg albumin (Sigma) and rotated for another 90 min. Beads were washed four times with 100 μl of C-100* buffer and boiled in SDS loading dye.
RNAi and assays for Nanog expression.
46C ES cells were transfected with pSuper-Esrrb-sh1, pSuper-Esrrb-sh2, or pSuper-control using Lipofectamine 2000 transfection reagent (Invitrogen). To measure the effect of Esrrb RNAi on the endogenous mRNA and protein levels of Nanog, Oct4, and Esrrb, transfected cells were selected with 1 μg/ml puromycin for 48 h, starting at 24 h posttransfection. RNA was isolated from these samples using Trizol (Invitrogen), and mRNA levels were measured by performing real-time quantitative PCR (qPCR) on an Opticon real-time PCR machine. Protein levels were measured by Western blot analyses using antibodies against Nanog (6), Oct4, Esrrb, and lamin B1 (Santa Cruz Biotechnology).
TNG-PS cells were transfected with pSuper-Esrrb-sh1, pSuper-Esrrb-sh2, or pSuper-control, and transfected cells were selected with puromycin for 48 h, starting at 24 h posttransfection. Green fluorescent protein (GFP) fluorescence of the TNG cells was measured by using a FACSCalibur flow cytometer (Becton Dickinson), as described previously (6).
For measuring the effect of Esrrb RNAi on expression from the Nanog promoter, pNanog-Luc containing a Nanog promoter fragment from −2.5 kb to +50 bp (11) and pRenilla-TK (Promega) were cotransfected with the pSuper constructs. Renilla luciferase assays were done 48 h posttransfection using the dual luciferase reporter system (Promega). The pNanog-Luc mutated Oct binding site (mOS) and its control were described previously (26). pNanog-Luc constructs with a mutant estrogen-related receptor response element (ERRE) contained the mutation GGT to AAC in the ERRE sequence TCTGGGTCA in the Nanog proximal promoter from −230 to +106, compared to the Nanog transcriptional start, and were tested 24 h posttransfection.
ChIP.
ChIP using formaldehyde cross-linking and Oct4 antibodies (sc-8628; Santa Cruz Biotechnology) or Esrrb antibodies (R&D Systems) was done on 46C ES cells, as described previously (2). Dual cross-linking ChIP using formaldehyde and di(N-succinimidyl) glutarate (DSG) was performed on 46C and ZBHTc4 ES cells, as described previously (24). qPCR analysis was performed using DNA Engine Opticon 2. Relative enrichments were calculated by comparing the ChIP efficiency of the region of interest to that of an unrelated region (Amylase). Primers used to amplify the Nanog genomic region are as follows: 5′ (−550 to −462), CACAGGCTCTTTCTTCAGACTTG and TCTTGCTTGCTCTTCACATTGG; Oct-Sox (−215 to −60), TCCCTCCCTCCCAGTCTG and CCTCCTACCCTACCCACCC; and 3′ (+929 to +988), GGTAGAACCAAGAGGCTGCT and CATCACAACACGCACCTGA. Primers used for Zfp42 are as follows: −283 to −117, TGCATCCTCTGCTTGTGTAA and CAGAGCTGTCCCCTTGTCT; Rest (−3216 to −3071), CTCCCCTGGACAATAGCTTC and CGTCCTTCATTTCCTCAGTG; Dppa3 (−1770 to −1550), GATCCAGCTGGTCTGAGCTA and GTGCAGGGATCATAGGAGTG; and Lefty1 (−1264 to −1060), AAGCTGCAGACTTCATTCCA and CGGGGGATAGATGAAGAAAC (21). Primers used for Amylase are as follows: CTCCTTGTACGGGTTGGT and AATGATGTGCACAGCTGAA.
EMSAs.
293T cells were transfected with pPyCAGIP derivatives (20) expressing the cDNAs of FLAG-Esrrb, Oct4, or Sox2. After 24 h, cells were washed and harvested in phosphate-buffered saline, resuspended in lysis buffer (50 mM HEPES, pH 7.9, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1× complete EDTA-free protease inhibitor; Roche) and rotated at 4°C for 20 min. After being microcentrifuged at 13,000 rpm for 10 min, supernatants were divided into aliquots and stored at −80°C. The electrophoretic mobility shift assay (EMSA) was carried out as described previously (7). The antibodies for the supershift were added after the initial incubation for a further 10 min as follows: 2 μg of anti-Oct4 (sc-9081), 2 μg of anti-Sox2 (sc-17320), or 1 μg of FLAG M2 antibody (Sigma).
Immunofluorescence.
46C ES cells were grown on coverslips coated with 0.1% gelatin and stained with antibodies using a standard protocol. In short, cells were fixed in 4% paraformaldehyde and incubated with antibodies against Nanog (6), Oct4 (N19; Santa Cruz Biotechnology), and Esrrb (R&D Systems). Secondary antibodies are from Dako (fluorescein isothiocyanate swine anti-rabbit antibody), Molecular Probes (Alexa Fluor 594 goat anti-mouse antibody) and Jackson ImmunoResearch Laboratories (fluorescein isothiocyanate rabbit anti-goat antibody). Images were taken with an Axio Imager (Zeiss).
RESULTS
The Oct4 protein interacts with Esrrb.
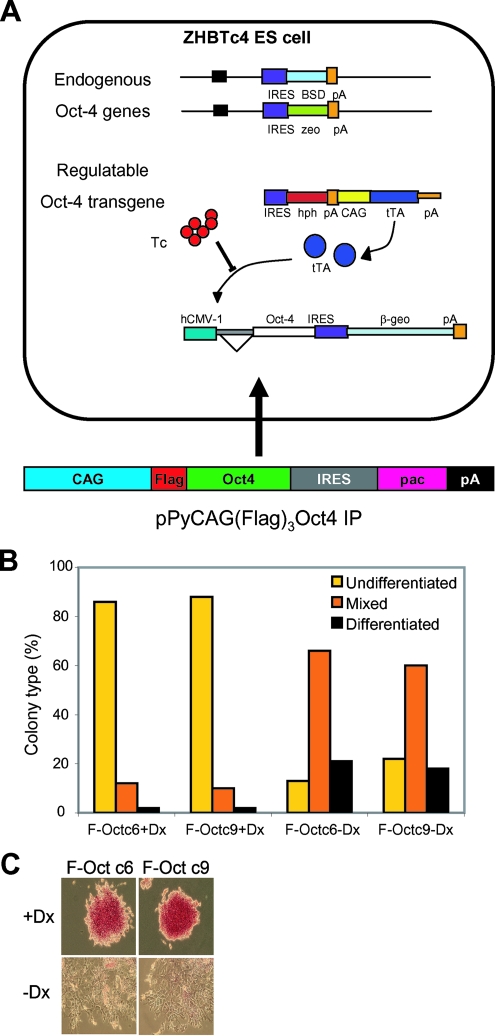
To identify the interaction partners of the Oct4 protein in ES cells, we constructed an ES cell line where, under self-renewing conditions, all Oct4 in the cell has an N-terminal triple FLAG tag (FLAG-Oct4). The parental ZHBTc4 ES cell line (23) has both Oct4 alleles disrupted, and the only Oct4 protein in the cell is transcribed from a doxycycline-suppressible transgene (Fig. 1A). ZHBTc4 cells were transfected with a construct in which the constitutive expression of FLAG-Oct4 is linked through an IRES to puromycin resistance. Simultaneously, doxycycline was added to the medium to repress the inducible Oct4 transgene expression (23). After 12 days of growth, colonies were picked and expanded into cell lines. All cell lines expressed a protein of the same relative molecular weight that reacted with an anti-FLAG antibody on Western blots (data not shown). As the Oct4 level must be tightly regulated to allow continued self-renewal (23), the survival of puromycin-resistant colonies indicates that the FLAG-Oct4 protein is functional. Two of these lines were further tested for their response to doxycycline treatment, and both underwent efficient differentiation at clonal density (Fig. 1B and C). The c6 cell line (F-Oct4 ES cells) was taken forward for biochemical analysis.
FIG. 1.
Construction and characterization of F-Oct ES cell lines. (A) In ZHBTc4 ES cells, both the Oct4 alleles have been replaced and Oct4 expression is directed from a doxycycline-suppressible transgene. F-Oct ES cell lines were derived from ZHBTc4 cells by transfection with linearized pPyCAG (FLAG)3 Oct4IP and the concomitant addition of doxycycline. (B) Clonal assays on two clones demonstrate that following the withdrawal of doxycycline, Oct4-induced differentiation occurs efficiently in representative F-Oct cell lines. Cells were plated at 600 cells per 10-cm dish in the presence or absence of 1 mM doxycycline, cultured for 6 days, and stained for alkaline phosphatase activity, and the differentiation status was determined. (C) Examples of colony morphologies in the presence (+Dx) and absence (−Dx) of doxycycline are shown.
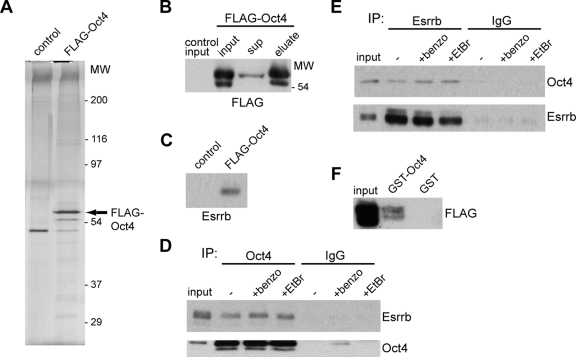
F-Oct4 ES cells were expanded, nuclear extracts were prepared, and the FLAG-Oct4 protein was purified using FLAG-affinity technology (see Materials and Methods). Silver stain analysis of a polyacrylamide gel containing the proteins purified from F-Oct4 extracts identified a major band and a minor band running just above the 54,000-molecular-weight marker (Fig. 2A). Both bands are recognized by a FLAG antibody and are not present in purifications from control extracts, suggesting that they represent the FLAG-Oct4 protein (Fig. 2B). No other major bands were observed, indicating that Oct4 does not purify as part of a major stoichiometric complex, despite the mild purification conditions used. Mass spectrometry analysis of two independent purifications identified the presence of Oct4 (10 unique peptides) and Esrrb (5 unique peptides) in F-Oct4 samples but not in control samples. Indeed, Esrrb could be detected by Western blot analysis in the F-Oct4 sample but not in the control (Fig. 2C). The interaction between Esrrb and Oct4 was independently verified by co-IP from extracts of a different ES cell line, 46C, using antibodies against endogenous Oct4 and Esrrb (Fig. 2D and E). Treatment of the extract with Benzonase nuclease or ethidium bromide did not affect the interaction (Fig. 2D and E), indicating that it is not indirectly mediated through DNA.
FIG. 2.
Oct4 interacts with Esrrb. (A) Silver-stained SDS-PAA gel of peptide-eluted FLAG-Oct4 versus control purification. The protein marker is shown as molecular weight (MW) in thousands. The band representing FLAG-Oct4 is indicated by the arrow. (B) Western blot analysis with anti-FLAG antibody on input, supernatant (sup), and elution of FLAG-Oct4 purification. (C) Western blot analysis with anti-Esrrb antibody on the eluted FLAG-Oct4 or control sample. (D, E) Co-IP experiments using antibodies against Oct4, Esrrb, or control immunoglobulin G (IgG) confirm the Oct4-Esrrb interaction. DNA independency of the interaction is shown by its insensitivity to Benzonase (benzo) or ethidium bromide (EtBr). (F) GST pulldown experiment using extracts from FLAG-Esrrb-transfected ES cells.
Moreover, the ability of bacterially expressed GST-Oct4 to pull down FLAG-Esrrb from transfected ES cells (Fig. 2F) indicates that posttranslational modification of Oct4 is not required for interaction between Oct4 and Esrrb.
Esrrb regulates expression of the Oct4 target gene Nanog.
Oct4 regulates expression of a cohort of target genes in ES cells, often acting in concert with Sox proteins (16). To determine whether the binding of Esrrb to Oct4 affected the regulation of gene expression by Oct4, we first examined expression of the Oct4 target gene Nanog. We depleted Esrrb by RNAi using two vectors that express different, previously reported Esrrb short hairpin RNAs (shRNAs) (16) and harbor a puromycin selection marker. After 2 days of puromycin selection, the levels of Nanog mRNA (Fig. 3A) and Nanog protein (Fig. 3B) were reduced in the 46C ES cells treated with either Esrrb shRNA vector compared to the levels of the control. Importantly, this specific depletion of Nanog occurred prior to any reduction in Oct4 expression (Fig. 3A and B) and prior to any morphological evidence of ES cell differentiation (Fig. 3C). This indicates that Esrrb shRNA-induced Nanog depletion is not a consequence of differentiation but occurs prior to differentiation. We also tested the effect of Esrrb depletion on TNG-PS ES cells which have the GFP open reading frame inserted at the start codon of one of the endogenous Nanog alleles (6). Transfection of either Esrrb shRNA vector decreased the mean GFP fluorescence of the population (Fig. 3D) after 2 days of puromycin selection, suggesting that depletion of Esrrb reduces transcription from the Nanog locus. To determine whether Esrrb affects the activity of the Nanog promoter, similar shRNA experiments were performed using a luciferase reporter under the control of a Nanog promoter fragment extending from −2.5 kb to +50 bp compared to the transcription start site. Esrrb shRNA vectors were cotransfected with the luciferase reporters, and luciferase activity was measured 2 days posttransfection. Figure 3E shows that Nanog promoter activity is strongly reduced with either Esrrb shRNA vector compared to activity with a control shRNA vector.
FIG. 3.
Esrrb regulates Nanog expression. (A) Real-time qPCR analysis shows downregulation of the mRNA levels of Nanog but not Oct4, following the shRNA-mediated knockdown of Esrrb in 46C ES cells by transfection of pSUPER plasmids expressing Esrrb shRNA1 or Esrrb shRNA2. RNA levels are compared to cells transfected with pSUPER expressing a control shRNA. Error bars represent the standard error of the mean (SEM) of three independent experiments. (B) Western blot analysis on total cell lysates confirms depletion at the protein level of Esrrb and Nanog, but not Oct4, upon shRNA-mediated knockdown of Esrrb. One of two independent experiments is shown. Lamin B1 is used as loading control. (C) Phase-contrast images of live 46C ES cell cultures 3 days after transfection of the indicated shRNA constructs. Scale bars represent 200 μm. (D) Fluorescence-activated cell sorter profiles of TNG-PS ES cells, in which the GFP open reading frame has been placed at the Nanog start codon. Knockdown of Esrrb with either of two shRNA constructs reduces GFP expression compared to that of the control construct. E14/T is an ES cell line lacking a GFP gene. One of two independent experiments is shown. (E) Luciferase reporter assays with a Nanog-promoter construct. The luciferase activity of the Nanog promoter (−2.5 kb to +50 bp), cotransfected with control shRNA plasmid, is arbitrarily set at 10 and compared to the luciferase activities in the presence of either of two Esrrb shRNA plasmids or of the pGL-Basic control vector. Error bars represent the SEM of three independent experiments.
Esrrb regulates Nanog expression using contacts with both Oct4 and a degenerate ERRE.
Oct4 contributes to the regulation of Nanog by binding to an Oct-Sox site (14, 26), located 166 to 180 bp upstream of the mapped transcription start site of the Nanog gene (4, 32). To investigate the relationship between the regulation of Nanog by Esrrb and that by Oct4, we performed ChIP experiments with antibodies for Oct4 and Esrrb in 46C ES cells and examined the precipitates for the presence of the Nanog promoter. A standard ChIP protocol using only formaldehyde as a cross-linking agent confirms that Oct4 binds in the vicinity of the Oct-Sox site (Fig. 4B). Using standard ChIP, we found no enrichment of Esrrb at the Nanog promoter (Fig. 4B), although the Esrrb protein is immunoprecipitated during the ChIP procedure (data not shown). Conventional formaldehyde-based ChIP methods efficiently detect protein-DNA interactions but may not detect the binding of Esrrb to the Nanog promoter if it is stabilized by protein-protein interactions. We therefore used a dual cross-linking ChIP method (XX-ChIP) that uses DSG prior to formaldehyde cross-linking (24). DSG has a longer spacer arm than formaldehyde and has been used to cross-link transcription factor protein-protein interactions on DNA (15). XX-ChIP indeed detects Esrrb at the Oct-Sox site within the proximal Nanog promoter (Fig. 4C). XX-ChIP using the Oct4 antibody also gives a specific enrichment of Oct4 at the Oct-Sox motif (Fig. 4C). Oct4 and Esrrb were also specifically enriched on the Oct-Sox site compared to the input and an immunoglobulin G control (data not shown). We conclude that Esrrb binds to the Nanog promoter in the vicinity of the Oct-Sox motif.
FIG. 4.
Esrrb binds to the Nanog promoter in an Oct4-dependent manner. (A) Outline of the Nanog genomic contig showing the amplicons (5′, Oct-Sox, and 3′) used in ChIP analysis and size markers in base pairs. (B) ChIP analysis of formaldehyde cross-linked 46C ES cells using antibodies against Oct4 and Esrrb. Relative enrichments of the Nanog amplicons are depicted as enrichments over that at an unrelated region (Amylase). Oct4 binds to the Oct-Sox element, but no significant enrichment for Esrrb can be detected at this region. Error bars represent the standard error of the mean (SEM) of two independent experiments. (C) ChIP analysis of dual cross-linked 46C ES cell chromatin shows enrichment of both Oct4 and Esrrb on the Oct-Sox element of the Nanog promoter. Error bars for Esrrb enrichments represent the SEM of two independent experiments. (D) Western blot analysis showing the complete depletion of Oct4 protein in ZHBTc4 ES cells after a 12-h treatment with 1 μg/ml doxycycline compared to that in untreated cells. Levels of Esrrb are not affected. Lamin B1 is used as a loading control. (E) Dual cross-linked chromatin from ZHBTc4 ES cells that were untreated (−Dox) or treated for 12 h with 1 μg/ml doxycycline (+Dox). Binding of Esrrb to the Oct-Sox element is no longer detected when Oct4 is absent. Error bars represent the standard error of the mean (SEM) of two independent experiments. (F) Luciferase assays with the wt Nanog promoter construct (Nanog-wt), with the mutated Oct binding site (Nanog-mOS) or with the empty vector pGL-Basic. Cotransfected shRNA plasmids are indicated. Error bars represent the SEM of three independent experiments.
To assess whether Esrrb binding to the Nanog promoter is dependent on Oct4, we made use of the ES cell line ZHBTc4 in which the endogenous Oct4 alleles are disrupted and in which Oct4 is expressed from a doxycycline-suppressible promoter (23). The addition of doxycycline for 12 h removes all Oct4 protein from the cell (Fig. 4D). At this time point, the level of Esrrb is unaffected (Fig. 4D). Esrrb XX-ChIP shows that Esrrb is no longer enriched at the Nanog promoter in the absence of Oct4 (Fig. 4E). To functionally test whether the maintenance of Nanog promoter activity by Esrrb is via Oct4, we tested Nanog promoter-luciferase constructs where the Oct4 binding site is mutated (Nanog mOS) (26). As expected, the Nanog mOS reporter is less active than the wild-type (wt) construct, although still clearly above the background (26) (Fig. 4F). Depletion of Esrrb does not further reduce the activity of Nanog mOS (Fig. 4F), suggesting that the effect of Esrrb on Nanog expression requires Oct4 binding to the Oct-Sox site in the Nanog promoter.
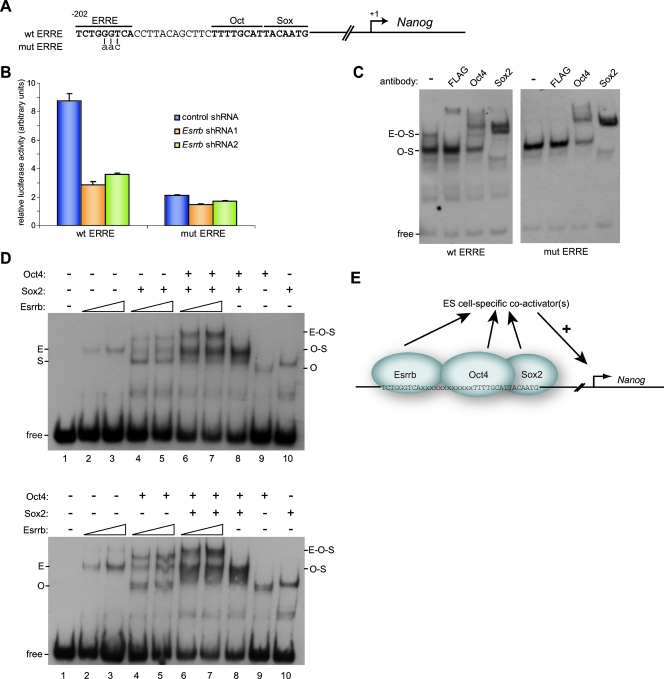
Estrogen-related receptors are thought to act via an ERRE. A consensus ERRE of tcaaGGttca (invariant positions in uppercase) was determined by SELEX and confirmed by in vivo studies (9, 29). Visual inspection of the Nanog promoter identified a degenerate ERRE (sequence TCTGGGTCA) 12 bp upstream of the Oct-Sox motif (Fig. 5A). This sequence is largely conserved in many mammalian species (26). To test the contribution of this putative ERRE to Nanog promoter activity, we mutated the core GGT in this motif into AAC (Fig. 5A) in the context of a Nanog promoter-luciferase construct. Nuclear magnetic resonance structural analysis of the Esrrb DNA binding domain in complex with DNA shows that these bases make a major contribution to DNA binding by Esrrb (10). This mutation strongly reduced Nanog promoter activity (Fig. 5B). Moreover, in contrast to the situation with the unmutated construct, the activity of this mutant could not be further reduced by Esrrb shRNA expression (Fig. 5B).
FIG. 5.
A degenerate ERRE binds Esrrb and is important for Nanog promoter activity. (A) Representation of the degenerate ERRE upstream of the Oct-Sox site in the Nanog promoter. The generated mutation is indicated. (B) Luciferase reporter assays with a wt Nanog promoter construct (wt ERRE) or mutated ERRE promoter construct (mut ERRE). Cotransfected shRNA plasmids are indicated. Error bars represent the standard error of the mean of three independent experiments. (C) Cell lysates from 293T cells cotransfected with FLAG-Esrrb, Oct4, and Sox2 expression plasmids were used in an EMSA with a 57-nucleotide Nanog promoter probe containing the Oct-Sox site sequence and a wt or mutated ERRE. Antibodies were added as indicated. The complex of Esrrb-Oct4-Sox2 (E-O-S) is indicated. (D) Cell lysates from 293T cells transfected with FLAG-Esrrb, Oct4, or Sox2 expression plasmids were subjected to EMSA using the wt Nanog promoter probe. (E) Model showing the enhancement of Esrrb binding to the Nanog promoter by Oct4 and Sox2 which positively affects Nanog promoter activity.
EMSA was employed to investigate the potential binding of Esrrb to the Nanog promoter. A 57-mer oligonucleotide corresponding to the sequence of the Nanog promoter that includes the putative ERRE and the Oct-Sox site was used. Lysate prepared from 293T cells that were cotransfected with FLAG-Esrrb, Oct4, and Sox2 expression plasmids caused a shift in migration of the probe into two complexes (Fig. 5C, left). The faster migrating complex could be supershifted by antibodies against Oct4 and Sox2 but not by an anti-FLAG antibody. In contrast, the slower migrating band was supershifted by all three antibodies, suggesting that the slower migrating complex is formed by binding of Esrrb, Sox2, and Oct4 (Fig. 5C, left, lanes 2 to 4). Using a second 57-mer oligonucleotide that has the GGT-to-AAC mutation in the putative ERRE, only the faster of the two complexes was observed, and this could be shifted by antibodies against Oct4 and Sox2 but not by anti-FLAG antibody (Fig. 5C, right). Therefore, we conclude that there is an ERRE upstream of the Oct-Sox site in the Nanog promoter that is essential for Esrrb binding and optimal Nanog promoter activity.
To determine whether binding of Esrrb to the Nanog promoter in vitro is dependent upon binding of Oct4-Sox2, EMSAs were performed by mixing cell lysates prepared from individual transfections of Oct4, Sox2, and Esrrb into 293T cells. The addition of the Esrrb lysate caused a weak probe shift (Fig. 5D, lanes 2 and 3), showing that Esrrb alone can bind the Nanog promoter probe. However, in the presence of Oct4 and Sox2, DNA complex formation by Esrrb was enhanced compared to DNA complex formation by Esrrb alone (Fig. 5D, compare lanes 2 and 3 to lanes 6 and 7). This effect was greatest with both Oct4 and Sox2 present, suggesting that an Oct4-Sox2 complex is required for this cooperative effect. Using different combinations of Esrrb, Oct4, and Sox2 lysates leads to the same conclusions (Fig. 5D, bottom). These data, together with the Esrrb ChIP experiments (Fig. 4C and E), suggest a model (Fig. 5E) in which the presence of the Oct4-Sox2 complex bound to the Oct-Sox site in the Nanog promoter strongly enhances the intrinsic capacity of Esrrb to bind to an ERRE located upstream of the Oct-Sox site.
Esrrb binds and regulates other Oct4 target genes.
To determine the generality of the association of Esrrb with Oct4 on Oct4 target genes, we investigate a number of Oct4 target genes with a characterized Oct-Sox DNA element (Fig. 6A), using the ES cell line ZHBTc4. These genes all had a putative ERRE at different distances from the Oct-Sox site and in different orientations (Fig. 6A). XX-ChIP analysis showed that Esrrb binds near the Oct-Sox element in the promoters of Zfp42 (Rex1) and Rest but not near the Oct-Sox site of Dppa3 and Lefty1 (Fig. 6B). Interestingly, removing Oct4 from the promoters by doxycycline treatment (Fig. 4D) prevented the detection of Esrrb at the Rest promoter, whereas Esrrb binding to the Zfp42 promoter was unaffected.
FIG. 6.
Esrrb binds and regulates other Oct4 target genes. (A) The sequences surrounding the Oct-Sox motifs in the regulatory elements of the Nanog, Zfp42, Rest, Dppa3, and Lefty1 genes are shown with the position and orientation of the putative ERREs indicated. The distance from the 3′ nucleotide of the shown Oct motif to the transcription start site is indicated. (B) ChIP with anti-Oct4 or anti-Esrrb antibodies on dual cross-linked chromatin isolated from ZHBTc4 cells that were either untreated (−Dx) or treated for 12 h with 1 μg/ml doxycycline (+Dx) to downregulate Oct4 expression. Enrichments over that at a negative control region (Amylase) are depicted; error bars represent standard error of the mean (SEM) of two independent experiments. (C) qPCR analysis of transcript levels in cells transfected with control or Esrrb shRNA. Error bars represent SEM of two independent experiments.
We next determined the contribution of Esrrb to the expression of Zfp42 and Rest by Esrrb knockdown. Expression of Esrrb shRNAs caused a reduction in Zfp42 mRNA expression, whereas Rest mRNA expression was unaffected (Fig. 6C). We conclude that Esrrb binds near the Oct-Sox sites of two other Oct4 targets, Zfp42 and Rest, but that only in the case of Zfp42 does this binding contribute to its expression in ES cells.
Esrrb protein levels correlate with Nanog levels in ES cell colonies.
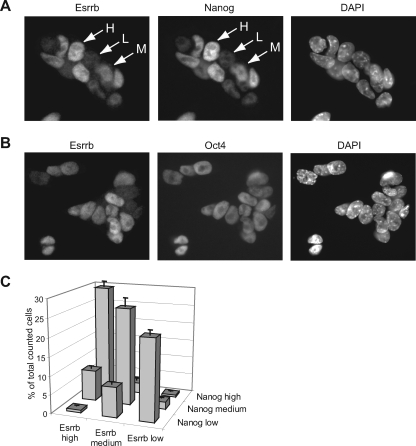
Oct4-positive (Oct4+) ES cell colonies express Nanog in a mosaic fashion (6, 12, 28). The cellular expression of Esrrb was examined by immunostaining ES cells with antibodies against Nanog or Esrrb. Interestingly, 46C ES cell colonies show a mosaic pattern of Esrrb expression within the Oct4+ population (Fig. 7A and B). Moreover, when cells are classified according to their relative high, medium, or low levels of Esrrb staining within ES cell colonies (Fig. 7A), there is a good correlation between the levels of expression of Nanog and those of Esrrb (Fig. 7C). This correlation between the cellular levels of Esrrb and Nanog, in combination with our Nanog gene regulation data, suggests that high Nanog expression in the cell may be facilitated by high Esrrb expression.
FIG. 7.
Esrrb protein levels in cells cofluctuate with Nanog protein levels. (A) Staining of ES cells with Esrrb antibody and Nanog antibody. Antibodies and DAPI staining are as indicated. Arrows point to cells that were assigned to have high (H), medium (M), or low (L) levels of Esrrb or Nanog. (B) Staining of ES cells with Esrrb antibody and Oct4 antibody. Antibodies and DAPI staining are as indicated. (C) Quantification of the percentage of cells that have high, medium, or low levels of Nanog or Esrrb. Error bars indicate the standard error of the mean of two independent experiments, in which 350 and 400 cells were assessed, respectively.
DISCUSSION
Using an Oct4 protein affinity strategy, we have identified Esrrb as a binding partner of the Oct4 protein. A previous report identified a number of other putative Oct4 interacting proteins but did not detect Esrrb (30). Our approach also identified a number of the reported interactors in our Oct4 sample, but these were often also present in the control sample. Our milder purification conditions may account for both the reproducible and verified identification of Esrrb as a specific Oct4 binding partner and the nonspecific binding of other reported interactors. Indeed, we find that Oct4-Esrrb interaction is sensitive to conditions with higher levels of salt (data not shown).
Members of the estrogen-related receptor family can bind to the palindromic 12-bp estrogen response element (25) or to the “extended half-site” 9-bp ERRE (9, 29). Either element can support Esrrb-mediated transcription (17, 33). Esrrb was also suggested to activate targets genes independently of a DNA element by binding to transcription factors, such as Sp1 (3). We show here, using ChIP and EMSAs, that Esrrb recruitment to the Nanog promoter requires both a degenerate, but conserved, ERRE and binding of Oct4 to the downstream Oct-Sox element. EMSAs (Fig. 5) confirm the previously reported synergistic binding of Oct4 and Sox2 to the Oct-Sox site (14), but also indicate that binding of Esrrb to the Nanog promoter oligonucleotides occurs cooperatively with binding of Oct4 and Sox2. This effect required binding of both Oct4 and Sox2.
We also provide evidence that recruitment of Esrrb to the Nanog promoter by Oct4 and the ERRE positively regulates Nanog expression. Depletion of Esrrb with shRNAs caused transcriptional downregulation of the endogenous Nanog gene and a Nanog promoter-reporter. Mutation of the ERRE also had a negative effect on reporter expression, underscoring the importance of the ERRE for the functional recruitment of Esrrb to the Nanog promoter.
Based upon the effect of mutations within the composite Oct-Sox site upon reporter gene expression directed by the proximal Nanog promoter, Oct4 has been suggested to be important for maintaining Nanog expression in ES cells (14, 26). However, Oct4 is not required to initiate Nanog expression in the preimplantation embryo, since Nanog transcripts (5) are present in Oct4−/− morulae and early blastocysts. This apparent discrepancy could be due, in part, to different requirements for establishment versus maintenance of Nanog expression. Our data on the Esrrb requirement for Nanog expression in ES cells suggest that one function of Oct4 binding to the Oct-Sox motif is that it facilitates Esrrb binding to the Nanog promoter, which in turn promotes Nanog transcription in ES cells.
Transcriptional regulation via transcription factor interactions in ES cell self-renewal.
Here we have provided evidence of a stem cell factor, Oct4, directing its physical interactor, Esrrb, to a target gene, Nanog, to positively regulate transcription. Gene regulation facilitated by complexes of individual transcription factors, like the Oct4-Esrrb complex, may be widespread in ES cells. Visual inspection of the sequences around the Oct-Sox sites of a number of Oct-Sox target genes for homology to the ERRE identified several potential Esrrb targets that were tested for regulation by Esrrb. Of these, ChIP analysis showed Esrrb to be detectable on Zfp42 and Rest but not Lefty1 or Dppa3. The Oct4-independent binding of Esrrb to the Zfp42 promoter may be due to the high match of the Zfp42 ERRE sequence to the consensus (8/9) (Fig. 6A), which may provide sufficient DNA binding affinity. The extreme proximity of the ERRE and Oct sites in Rest coupled with the low match to the ERRE consensus (6/9) could underlie the Oct4-dependent ChIP of Esrrb at Rest. However, the ERRE in Nanog is further removed from the Oct-Sox site and is a better match to the consensus (7/9) than that in Rest, so there is no obvious common feature that can explain the Oct4-dependent binding of Esrrb to each of these sequences. There is also no clear reason why the remaining two Oct-Sox targets do not bind Esrrb. A low match to the consensus could explain the lack of binding to Dppa3 (6/9). However, the consensus match for Lefty1 is the same as that for Nanog (7/9), suggesting that relative spatial disposition and/or distance could play a role. Further experimentation will be required to more deeply understand the relationship of Oct4 and Esrrb binding to DNA and how this affects gene regulation.
Nanog is expressed mosaically within the Oct4+ populations in ES cell cultures (6, 12, 28). Moreover, Nanog levels fluctuate in ES cell cultures such that cells expressing a low level or no Nanog can reexpress a high level of Nanog. However, lowered Nanog expression predisposes cells toward differentiation, without marking a commitment event (6). Here we show that the mosaic patterns of Esrrb and Nanog expression in ES cell colonies largely overlap. We also show that Esrrb positively regulates Nanog expression. As Nanog has been reported to positively regulate Esrrb expression (16), Esrrb and Nanog may both act to reinforce expression of the reciprocal gene through a positive feedback loop. How this leads to mosaic and cofluctuating levels of both proteins remains to be determined. Oct4 is not obviously mosaic and appears not to fluctuate, suggesting that it is not a determining factor of fluctuations in Nanog and Esrrb in ES cells. As Nanog levels and, by implication, Esrrb levels regulate the self-renewal efficiency of ES cells, unraveling this regulatory mechanism will be important for a fuller understanding of ES cell self-renewal and the maintenance of pluripotency.
Acknowledgments
We thank Hitoshi Niwa for ZHBTc4 cells, Austin Cooney for the 2.5-kb Nanog promoter-luciferase construct, Paul Robson for the Nanog mOS luciferase and control Nanog luciferase constructs, Austin Smith for 46C ES cells, Rodrigo Osorno for technical assistance, and Frank Grosveld for critically reading the manuscript and for helpful suggestions.
This work was supported by an NWO Vidi grant to the R.P. laboratory and by the Medical Research Council and the Biotechnological and Biological Sciences Research Councils of the United Kingdom, the Wellcome Trust, and the Juvenile Diabetes Research Foundation.
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Ambrosetti, D. C., C. Basilico, and L. Dailey. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 176321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, L. A., T. I. Lee, M. F. Cole, S. E. Johnstone, S. S. Levine, J. P. Zucker, M. G. Guenther, R. M. Kumar, H. L. Murray, R. G. Jenner, D. K. Gifford, D. A. Melton, R. Jaenisch, and R. A. Young. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122947-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castet, A., A. Herledan, S. Bonnet, S. Jalaguier, J. M. Vanacker, and V. Cavailles. 2006. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol. Endocrinol. 201035-1047. [DOI] [PubMed] [Google Scholar]
- 4.Chambers, I. 2005. Mechanisms and factors in embryonic stem cell renewal. Rend. Fis. Acc. Lincei 1683-97. [Google Scholar]
- 5.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113643-655. [DOI] [PubMed] [Google Scholar]
- 6.Chambers, I., J. Silva, D. Colby, J. Nichols, B. Nijmeijer, M. Robertson, J. Vrana, K. Jones, L. Grotewold, and A. Smith. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 4501230-1234. [DOI] [PubMed] [Google Scholar]
- 7.Chew, J.-L., Y.-H. Loh, W. Zhang, X. Chen, W.-L. Tam, L.-S. Yeap, P. Li, Y.-S. Ang, B. Lim, P. Robson, and H.-H. Ng. 2005. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 256031-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour, C. R., B. J. Wilson, J. M. Huss, D. P. Kelly, W. A. Alaynick, M. Downes, R. M. Evans, M. Blanchette, and V. Giguere. 2007. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab. 5345-356. [DOI] [PubMed] [Google Scholar]
- 10.Gearhart, M. D., S. M. Holmbeck, R. M. Evans, H. J. Dyson, and P. E. Wright. 2003. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J. Mol. Biol. 327819-832. [DOI] [PubMed] [Google Scholar]
- 11.Gu, P., D. LeMenuet, A. C.-K. Chung, M. Mancini, D. A. Wheeler, and A. J. Cooney. 2005. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell. Biol. 258507-8519. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 12.Hatano, S. Y., M. Tada, H. Kimura, S. Yamaguchi, T. Kono, T. Nakano, H. Suemori, N. Nakatsuji, and T. Tada. 2005. Pluripotential competence of cells associated with Nanog activity. Mech. Dev. 12267-79. [DOI] [PubMed] [Google Scholar]
- 13.Ivanova, N., R. Dobrin, R. Lu, I. Kotenko, J. Levorse, C. DeCoste, X. Schafer, Y. Lun, and I. R. Lemischka. 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442533-538. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda, T., M. Tada, H. Kubota, H. Kimura, S.-Y. Hatano, H. Suemori, N. Nakatsuji, and T. Tada. 2005. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 252475-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, T., A. E. Tee, A. Porro, S. A. Smith, T. Dwarte, P. Y. Liu, N. Iraci, E. Sekyere, M. Haber, M. D. Norris, D. Diolaiti, G. Della Valle, G. Perini, and G. M. Marshall. 2007. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis. Proc. Natl. Acad. Sci. USA. 10418682-18687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh, Y. H., Q. Wu, J. L. Chew, V. B. Vega, W. Zhang, X. Chen, G. Bourque, J. George, B. Leong, J. Liu, K. Y. Wong, K. W. Sung, C. W. Lee, X. D. Zhao, K. P. Chiu, L. Lipovich, V. A. Kuznetsov, P. Robson, L. W. Stanton, C. L. Wei, Y. Ruan, B. Lim, and H. H. Ng. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38431-440. [DOI] [PubMed] [Google Scholar]
- 17.Lu, D., Y. Kiriyama, K. Y. Lee, and V. Giguere. 2001. Transcriptional regulation of the estrogen-inducible pS2 breast cancer marker gene by the ERR family of orphan nuclear receptors. Cancer Res. 616755-6761. [PubMed] [Google Scholar]
- 18.Masui, S., Y. Nakatake, Y. Toyooka, D. Shimosato, R. Yagi, K. Takahashi, H. Okochi, A. Okuda, R. Matoba, A. A. Sharov, M. S. Ko, and H. Niwa. 2007. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9625-635. [DOI] [PubMed] [Google Scholar]
- 19.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113631-642. [DOI] [PubMed] [Google Scholar]
- 20.Mullin, N. P., A. Yates, A. J. Rowe, B. Nijmeijer, D. Colby, P. N. Barlow, M. D. Walkinshaw, and I. Chambers. 2008. The pluripotency rheostat Nanog functions as a dimer. Biochem. J. 411227-231. [DOI] [PubMed] [Google Scholar]
- 21.Nakatake, Y., N. Fukui, Y. Iwamatsu, S. Masui, K. Takahashi, R. Yagi, K. Yagi, J.-I. Miyazaki, R. Matoba, M. S. H. Ko, and H. Niwa. 2006. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol. Cell. Biol. 267772-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa, H. 2007. How is pluripotency determined and maintained? Development 134635-646. [DOI] [PubMed] [Google Scholar]
- 23.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24372-376. [DOI] [PubMed] [Google Scholar]
- 24.Nowak, D. E., B. Tian, and A. R. Brasier. 2005. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. BioTechniques 39715-725. [DOI] [PubMed] [Google Scholar]
- 25.Pettersson, K., K. Svensson, R. Mattsson, B. Carlsson, R. Ohlsson, and A. Berkenstam. 1996. Expression of a novel member of estrogen response element-binding nuclear receptors is restricted to the early stages of chorion formation during mouse embryogenesis. Mech. Dev. 54211-223. [DOI] [PubMed] [Google Scholar]
- 26.Rodda, D. J., J.-L. Chew, L.-H. Lim, Y.-H. Loh, B. Wang, H.-H. Ng, and P. Robson. 2005. Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 28024731-24737. [DOI] [PubMed] [Google Scholar]
- 27.Sánchez, C., I. Sánchez, J. A. Demmers, P. Rodriguez, J. Strouboulis, and M. Vidal. 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell. Proteomics 6820-834. [DOI] [PubMed] [Google Scholar]
- 28.Singh, A. M., T. Hamazaki, K. E. Hankowski, and N. Terada. 2007. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 252534-2542. [DOI] [PubMed] [Google Scholar]
- 29.Sladek, R., J.-A. Bader, and V. Giguere. 1997. The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell. Biol. 175400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, J., S. Rao, J. Chu, X. Shen, D. N. Levasseur, T. W. Theunissen, and S. H. Orkin. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444364-368. [DOI] [PubMed] [Google Scholar]
- 31.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379466-469. [DOI] [PubMed] [Google Scholar]
- 32.Wu, D. Y., and Z. Yao. 2005. Isolation and characterization of the murine Nanog gene promoter. Cell Res. 15317-324. [DOI] [PubMed] [Google Scholar]
- 33.Xie, W., H. Hong, N. N. Yang, R. J. Lin, C. M. Simon, M. R. Stallcup, and R. M. Evans. 1999. Constitutive activation of transcription and binding of coactivator by estrogen-related receptors 1 and 2. Mol. Endocrinol. 132151-2162. [DOI] [PubMed] [Google Scholar]
- 34.Ying, Q. L., J. Nichols, I. Chambers, and A. Smith. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115281-292. [DOI] [PubMed] [Google Scholar]
- 35.Ying, Q. L., M. Stavridis, D. Griffiths, M. Li, and A. Smith. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21183-186. [DOI] [PubMed] [Google Scholar]