Abstract
High-throughput sequencing has rapidly gained popularity for transcriptome analysis in mammalian cells because of its ability to generate digital and quantitative information on annotated genes and to detect transcripts and mRNA isoforms. Here, we described a double-random priming method for deep sequencing to profile double poly(A)-selected RNA from LNCaP cells before and after androgen stimulation. From ≈20 million sequence tags, we uncovered 71% of annotated genes and identified hormone-regulated gene expression events that are highly correlated with quantitative real time PCR measurement. A fraction of the sequence tags were mapped to constitutive and alternative splicing events to detect known and new mRNA isoforms expressed in the cell. Finally, curve fitting was used to estimate the number of tags necessary to reach a “saturating” discovery rate among individual applications. This study provides a general guide for analysis of gene expression and alternative splicing by deep sequencing.
Keywords: alternative splicing, androgen-regulated gene expression in prostate cancer cells, curve regression, high-throughput sequencing
Microarray-based approaches, especially unbiased tiling arrays, suggest that up to 80% of the genome may be transcribed to produce a huge number of uncharacterized transcripts relative to current gene annotation (1–4). In contrast, recent transcriptome analysis by deep sequencing indicates that the vast majority of expressed transcripts in mammalian tissues and cell lines are confined to annotated genes and exons (5, 6). Although microarray-based approaches suffer from a great degree of uncertainty in relating detected hybridization signals to defined transcripts, sequencing-based approaches tend to be overwhelmed by abundant transcripts in the cell. Construction of “normalized” libraries for deep sequencing might facilitate the discovery of low abundance transcripts, many of which may act as noncoding, regulatory RNA in mammalian cells.
An advantage of transcriptome analysis by deep sequencing is the ability to detect structural variation of individual transcripts. It is well known that different transcripts from the same genes may be generated by differential promoter usage, heterogeneous transcriptional start sites and alternative 3′ end formation (2). A recent Pol II ChIP-chip study indicates that protein-coding genes may have an average of 3 to 5 promoters in both the mouse and human genomes (7). Further adding to the diversity in the transcriptome is alternative RNA processing of most protein-coding genes as a consequence of alternative 5′ and 3′ splice site choices, exon inclusion/skipping, intron retention, and combinatorial use of alternative exons (8). It is estimated that up to 74% of human genes undergo alternative splicing, which is believed to contribute to the complexity of the proteome in mammalian cells (9).
It is striking to note that individual laboratories now have the capacity to generate sequenced tags that are on the same order of sequenced mRNA/ESTs in publicly available databases (≈160K mouse mRNAs from RIKEN (10) and ≈30M ESTs in dbEST) accumulated over decades. However, despite recent reports on transcriptome analysis by deep sequencing (5, 6, 11), a range of practical issues remain to be addressed: How many annotated genes are detectable in a single cell type, what is the number of tags that is necessary for quantitative analysis of differentially regulated genes under different experimental conditions, to what extent can existing mRNA isoforms be detected, and how can one quantify alternative splicing by using a single or combination of existing technologies?
In this report, we attempted to address these issues on an androgen-sensitive prostate cancer cell model. Using a double-random priming approach capable of generating strand-specific information, we sequenced poly(A)+ RNA from mock-treated or androgen-stimulated LNCaP cells on the Illumina 1G Genome Analyzer. Analysis of ≈10 million sequence tags generated from both control and hormone-treated cells suggests that this tag density is sufficient for quantitative analysis of gene expression. We were also able to detect a large fraction of tags corresponding to annotated alternative exons, with a subset of the tags matching known and detecting new splice junctions; however, the current tag density is insufficient to deduce quantitative differences among most detected mRNA isoforms. Based on this information, a computational model based on curve fitting was used to estimate the tag density needed for optimal detection of regulated gene expression and alternative splicing.
Results and Discussion
Transcriptome Analysis by Double-Random Priming.
Typical RNAseq procedures used by several published studies involve random priming to convert poly(A)+ mRNA to double-stranded cDNA followed by linker ligation (5, 6, 12, 13). To simplify library construction for deep sequencing, we devised a procedure based on double-random priming and solid phase selection (Fig. 1). In this procedure, the first random primer (octamer linked to the sequencing primer P1) was used to prime double poly(A)-selected RNA from LNCaP cells. A variation of this is to use oligo(dT) linked to the sequencing primer for analysis of total RNA without poly(A) selection. The first primer also carries a biotin moiety at the 5′ end, which allows transfer of extended cDNA to streptavidin beads. The second random primer linked to the other sequencing primer (P2) was next used to prime the cDNA on the streptavidin beads. After extensive washes, potential P2 dimers were eliminated and the second random primed products were released from the beads by heat, leaving behind unused P1 primer, P1-extended cDNA, and potential P1 dimers. The released products are PCR-amplified, gel purified to enrich for amplicons in the size range of 100–300 nt, quantified, and subjected to sequencing from the P1 primer on the Illumina/Solexa flowcell.
Fig. 1.
The double random priming method for deep sequencing. The first biotinylated random primer consists of the sequencing primer P1 at the 5′ end and a random octamer at the 3′ end. Products of the first random priming reaction were selected on streptavidin beads (blue eclipse) followed by the second random priming reaction on the solid phase with a random octamer carrying the sequencing primer P2. After extensive washes to remove free primers and primer dimmers, the second random priming products were released from beads by heat, which were then PCR-amplified, gel-purified, and subjected to sequencing from the P1 primer.
This procedure has a number of useful features. First, it provides strand-specific information. Second, sequencing a short region right after the first random priming reaction avoids cDNA artifacts resulting from extension by the hairpins formed after the first strand synthesis (14), which may account for artifactual “antisense transcripts” seen in previous large-scale mRNA sequencing and tiling analysis (1, 15). Third, the built-in random primer region retains the molecular memory for originally primed products allowing computational elimination of sequenced tags amplified by PCR, because all PCR products from the same initial amplicon will have identical sequences in the randomized region. This strategy permits the use of PCR amplification without distorting the representation of the transcriptome, a feature critical for quantitative analysis on a small population of cells.
Global Statistics of Gene Expression.
We obtained ≈10 million 36-nt sequence tags from mock- and androgen-treated LNCaP cells, respectively. To use the longest possible read for mapping the tags to specific transcripts and to splice junctions, we only removed the first nucleotide from the random primer region because it contributes the least to the random priming reaction (Fig. S1). By allowing 2 base mismatches, we were able to uniquely align ≈21% of the tags to the human genome (Fig. 2A). This is lower than previously published results (5, 6, 11), but is not due to contamination in our purified poly(A) RNA by transcripts from repeat regions, including rRNA, in the human genome, which together constitute <1% of total sequenced tags (Fig. 2A). An analysis of nucleotide frequencies of mapped tags suggests a degree of bias during random priming for relatively high GC-content, especially at positions +5 to +7 (Fig. S1A). The uniquely mapped tag set also allowed us to determine the sequencing error at each position, indicating a high error rate at both the random priming region and toward the end of the sequenced tags (Fig. S1B). By including tags that were mapped up to 5 positions in the human genome and/or removing the first 4 nt and last 3 nt, we were able to progressively increase the number of “mappable” tags (Table S1). We nevertheless elected to use the longest tags with most stringent alignment for downstream analyses.
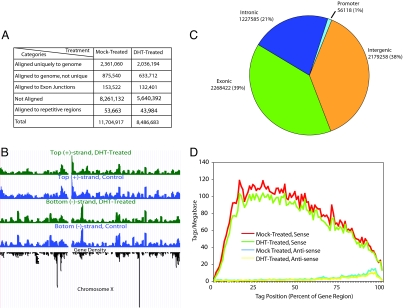
Fig. 2.
Global mapping of sequence tags. (A) Summary of genomic mapping results, allowing 2 mismatches in 35 nt. For comparison, additional mapping results that include tags that hit up to 5 positions in the genome or with tags after removal of the first 4 nt and last 3 nt are shown in Table S1. Sequence tags mapped to splice junctions include known junctions and junctions determined in this study. (B) Transcription from top (+) and bottom (−) strands of human chromosome X. The data showed high reproducibility with high (>0.7) Pearson correlation coefficients within the same strands and low (<0.04) correlation between different strands (see Table S2). (C) Genomic distribution of sequence tags in exons, introns, promoters (3 kb from transcription start sites), and intergenic regions. (D) Sense and antisense transcripts. Sequence tags corresponding to both sense and antisense transcripts were color-coded and displayed on a composite mRNA map with the x axis showing the tag position (% of spliced mRNA region) and the y axis showing the tag density (tags per megabase).
Because of the strand specificity of our method, we were able to unambiguously assign tags to either the top (+) or the bottom (−) strand of the human genome according to the genome orientation (Fig. 2B). The tag distribution is highly reproducible as evidenced by plotting the tags along human chromosomes under two biological conditions (Fig. 2B). Furthermore, high Pearson correlation coefficients within the same strand under different treatment conditions and low Pearson correlation coefficients between different strands under the same conditions indicate a high degree of data reproducibility and a minimal impact of global gene expression by the hormone treatment (Table S2).
The genomic distribution of the mapped tags according to current gene annotation shows that most tags are confined to protein coding genes, although we detected a large number of tags in intronic regions (Fig. 2C). This profile is distinct from other published studies based on the Solexa platform, but comparable to those observed by 454 sequencing (5, 6, 16). The exact reason(s) for this discrepancy is presently unclear. We also detect a sizable fraction of tags in intergenic regions, which correspond to potentially new coding and noncoding transcripts but little in gene promoters, as expected (Fig. 2C).
Consistent with the observations made by large-scale full-length cDNA sequencing (10) and tiling array studies (1), we also detected a sizable number of antisense transcripts and aligned both sense and antisense transcripts on the normalized gene model (Fig. 2D). For sense transcripts, the tag density is low at the 5′ end, likely due to heterogeneous transcription start and to size selection of amplicons for sequencing, which removes small amplicons (<100 nt) from the 5′ end. Interestingly, the tag density for sense transcripts declines toward the 3′ end. Although the exact reason for this is presently unclear, such steady decline is distinct from that at the 5′ end, indicating that the pattern cannot simply be explained by size-selection or alternative 3′ end formation. In contrast, most tags aligned to antisense transcripts were detected at the 3′ end of protein-coding genes (Fig. 2D). This profile is consistent with the binding of many common transcription factors, including Pol II, at the 3′ end of many genes (17). Because antisense transcripts are thought to provide some regulatory function to sense transcription, their termination toward the 5′ end may result from competition with sense transcription as suggested by the polymerase collision model (18). It is yet to be determined whether any detected sense/antisense pairs are subjected to androgen regulation.
Quantitative Detection of Androgen-Responsive Genes.
The high density of tags mapped onto annotated genes provides both qualitative and quantitative measures of transcription in response to the hormonal signal. We detected the expression of 71% annotated genes in LNCaP cells (Table 1). To determine hormone-regulated genes in this widely used prostate cancer cellular model, we enumerated the number of tags mapped to exons in individual transcripts before and after DHT stimulation. To identify hormone-regulated genes, we compared the number of tags mapped to specific transcripts to the total number of tags mapped to all other transcripts, using χ2 statistics (Fig. 3A). At the cutoff of P < 0.01, we identified 359 genes (red dots in Fig. 3A) that were differentially up- or down-regulated by hormone treatment (Table S3). Real time RT-PCR validated a sizable set of androgen-responsive genes, demonstrating a high concordance (R2 = 0.92) between the digital information generated by deep sequencing and the quantitative measurement by qPCR (Fig. 3B and Table S4).
Table 1.
Summary of curve-fitting results and predictions
| R2(y = axb) | R2 (y = aebx) | a | b | Features in database | Current Density: (4,397,254) |
5% Discovery Rate |
1% Discovery Rate |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Features detected | Yield, % | Estimated mappable tags required | Estimated features detected | Yield, % | Estimated mappable tags required | Estimated features detected | Yield, % | ||||||
| Transcripts detected | 0.9661 | 0.9005 | 294,288,261 | −1.6074 | 17,478 | 12,431 | 71 | 1,196,539 | 11,630 | 67 | 3,256,610 | 12,345 | 71 |
| Differentially expressed transcripts | 0.9045 | 0.8532 | 4,131,205,578 | −1.6192 | 359 | 5,526,079 | 366 | 14,931,442 | 560 | ||||
| Constitutive exons | 0.9965 | 0.9266 | 2,050,312,589 | −1.6247 | 191,791 | 55,789 | 29 | 3,404,656 | 51,890 | 27 | 9,168,122 | 56,600 | 30 |
| Alternative exons, mRNA/EST support | 0.9810 | 0.9206 | 1,609,879,781 | −1.6033 | 18,683 | 3,631 | 19 | 3,580,965 | 3,495 | 19 | 9,771,601 | 3,670 | 20 |
| ACEscan[+] exons | 0.9393 | 0.8637 | 835,763,570 | −1.5633 | 4,486 | 1,030 | 23 | 3,463,185 | 965 | 22 | 9,695,935 | 1,040 | 23 |
| Constitutive junctions (CE) | 0.9305 | 0.9851 | 10.6522 | −1.91838 × 10−6 | 142,084 | 29,384 | 21 | 2,794,799 | 22,340 | 16 | 3,633,754 | 29,340 | 21 |
| Alternative splice junctions, EST/mRNA support (AS) | 0.9269 | 0.9836 | 9.4778 | −1.86229 × 10−6 | 51,698 | 8,328 | 16 | 2,816,263 | 6,235 | 12 | 3,680,490 | 8,315 | 16 |
| ACEscan[+] junctions (ACE) | 0.9351 | 0.9837 | 9.0100 | −1.8563 × 10−6 | 10,687 | 1,678 | 16 | 2,798,077 | 1,270 | 12 | 3,665,091 | 1,675 | 16 |
Fig. 3.
Digital analysis of androgen-regulated gene expression in LNCaP cells. (A) Scatter plot of gene expression in mock-treated and DHT-induced cells. Differential expressed genes were labeled red based on χ2 (P < 0.01). (B) Comparison of fold changes determined by sequencing and by quantitative measurement with real time PCR. (C) Comparison with 5 published microarray datasets in LNCaP cells. The currently determined androgen-regulated genes showed 25% overlap with at least one published microarray study as indicated by color-coded sections in the pie-chart. Specifically, 218 genes showed no overlap; 87 overlapped with 1 report; 30 with 2; 10 with 3; 9 with 4, and only 1 gene was common with all 5 published reports. Detailed comparisons of individual genes identified in the current and published studies were summarized in Table S6. (D) Curve fitting the change in the number of new features detected relative to increasing tag densities. Dashed line indicated exponential curve fit; solid line indicated power curve fit. R2 coefficients for each fitted curve were displayed in Table 1. The graph indicated that as the tag density increased the rate of identification of additional transcripts (blue) and DHT-induced transcripts (orange) decreased. The horizontal lines indicated where the discovery rate drops below 5% (red) and 1% (green).
Gene Ontology (GO) analysis indicated that the hormone up-regulated genes identified in the current study were enriched in categories involved in cellular signaling and nuclear functions (Table S5). Similarly to ref. 19, we compared our dataset with those derived from published microarray-based studies, we found that 25% of hormone responsive genes overlapped with at least one published profile (Fig. 3C). Although all pairwise comparisons of our differentially regulated genes showed statistically significant overlap (P < 0.01) with sets published in refs. 20–24, the degree of overlap varies considerably (Table S6), which likely results from a combination of different statistical cutoffs, different treatment conditions, variable biological response of the same cell type cultured in individual laboratories, and other experimental differences.
To estimate the number of tags required for detecting specific genomic features, in this case, the total number of annotated genes and differentially expressed genes, we developed an approach based on power or exponential curve fitting to extrapolate the optimal tag density for detection of individual genomic features (see Materials and Methods). Our calculations, which take into account the number of features observed at increasing tag densities, suggest that ≈1.2 million and ≈5.5 million mappable tags would be needed to reach a level where further increase in tag density will not yield >5% additional discovery of annotated genes and DHT-responsive genes, respectively. These numbers increase to ≈3.3 million and 14.9 million tags for annotated genes and DHT-responsive genes, respectively, if the threshold is set to 1% (Fig. 3D and Table 1). Although regulated gene expression induced by external signals varies among different cell types, our method provides an estimate for the tag density needed for both qualitative (expressed transcripts) and quantitative (differentially expressed transcripts) analyses of gene expression.
Strategies to Identify Alternative Exons and Splice Junctions.
It is striking that a large fraction (≈70%) of sequence tags could not be mapped at our stringent criteria to the reference human genome, a certain percentage of which likely corresponds to splice junctions created by constitutive and alternative mRNA splicing. Although several published deep-sequencing studies have documented the ability of using short sequence reads to identity splice junctions (5, 6, 12), it is unclear to what extent the current sequencing technologies are able to uncover mRNA isoforms and whether it is practical to comprehensively profile alternative splicing by deep sequencing. We attempted to address these questions with the data generated from LNCaP cells, using two complementary approaches.
Our first approach was to construct an exon-body database (EBDB) by identifying all exons that are flanked by canonical splice signals (GT-AG, AT-AC, GC-AG), which were further classified into constitutive exons (CE), alternative exons that are supported by the existing mRNA/EST information (AE), and alternative conserved exons (ACE) identified by the ACEscan algorithm (25). Mapping of the sequence tags to individual exons revealed 19 to 29% of exons in each class were detectable at current tag density (Table 1). Curve-fitting analysis suggests that our current density is approaching the predicted threshold such that no >1% additional discovery can be achieved with further increase in tag density (Fig. 4A and Table 1). This is likely related to variation in transcript abundance, because low abundance transcripts may not have sufficient tag densities to detect all their individual exons.
Fig. 4.
Curve fitting the change in the number of exons and splice junctions detected against increasing tag densities. Dashed line indicated exponential curve; solid line indicated power curve. R2 coefficients for each fitted curves were displayed in Table 1. (A) Decline in the rate of identifying additional exons as a function of increasing tag density. (B) Decline in the rate of identifying additional splice junctions as a function of increasing tag density. The horizontal lines in both panels indicate where discovery rate drops below 5% (red) and 1% (green).
Complementary to the exon body approach, we also created a splice junction database for constitutive and alternative splicing events supported by the existing mRNA/ESTs and alternative conserved splicing events identified by ACEscan (25). This allowed us to assign a fraction of previously unaligned tags to known splice junctions. To ensure accuracy, we conservatively required that at least two independent tags mapped to the same junction with at least 4 nt across the exon-exon junction. The rationale for requiring 4 nt overlap was based on examining the fraction of the correct exon-exon junction that shared identical sequence (thus “indistinguishable”) with the downstream intronic bases or the downstream exon junction as a function of increasing nucleotides across the junction (see the diagram in Fig. S2). If we allow only 1 nt across the splice junction, for instance, ≈46% of splice junctions were indistinguishable. The fraction of indistinguishable junctions decreases to only 1% if we require 4 nt across splice junctions. Using this strategy, we detected 16 to 21% of known splice junctions (constitutive and alternative) at the current tag density and curve-fitting analysis suggests that our current tag density is already at the predicted threshold (1%) (Fig. 4B, Table 1).
In an attempt to identify alternative splicing events, we used the simplest mode of alternative splicing (exon skipping) to construct a hypothetical exon-junction database (EJDB) by piecing together the 35 bases at the 3′ end of each internal exon to the 35 bases at the 5′ end of individual downstream exons. We next aligned the sequence tags to the EJDB, identifying 724 + 651 = 1,375 junctions, approximately half (724) of which span only constitutive exons (Table 2). Although this is by no means a comprehensive survey of new alternative splicing events because exon skipping is only one of multiple alternative splicing modes, the result indicates that the vast majority of alternative splicing events (8) from measurable abundant gene transcripts has already been covered by the existing mRNA/EST databases (Table 2).
Table 2.
Detection of known and novel splice junctions
| Exons skipped | EST-verified |
Novel |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CE |
AS |
ACE |
CE |
AS |
ACE |
|||||||
| Database size | Junctions detected | Database size | Junctions detected | Database size | Junctions detected | Database size | Detected | Database size | Detected | Database size | Detected | |
| 0 | 139,326 | 29,104 | 35,938 | 6,727 | 8,023 | 1,297 | 4,153 | 25 | 2,252 | 19 | 397 | 4 |
| 1 | 1,686 | 222 | 11,932 | 1,390 | 1,831 | 319 | 115,959 | 361 | 34,823 | 124 | 9,194 | 51 |
| 2 | 698 | 45 | 2,365 | 157 | 474 | 41 | 97,324 | 99 | 47,846 | 116 | 12,379 | 30 |
| 3 | 202 | 9 | 794 | 36 | 174 | 14 | 82,452 | 67 | 49,820 | 59 | 13,929 | 17 |
| 4 | 75 | 1 | 311 | 7 | 77 | 4 | 70,290 | 34 | 48,917 | 57 | 14,813 | 18 |
| 5 | 34 | 1 | 138 | 5 | 35 | 2 | 60,272 | 17 | 46,815 | 46 | 15,211 | 13 |
| 6 | 17 | 1 | 68 | 2 | 16 | 0 | 51,977 | 22 | 44,125 | 26 | 15,284 | 7 |
| 7 | 8 | 0 | 47 | 1 | 13 | 0 | 44,982 | 16 | 41,161 | 30 | 15,074 | 9 |
| 8 | 8 | 0 | 34 | 1 | 10 | 0 | 39,108 | 13 | 38,085 | 20 | 14,685 | 4 |
| 9 | 6 | 0 | 14 | 0 | 7 | 0 | 34,069 | 14 | 35,128 | 24 | 14,152 | 12 |
| 10 | 2 | 0 | 10 | 1 | 2 | 0 | 29,801 | 8 | 32,252 | 15 | 13,526 | 4 |
| Σ | 142,084 | 29,384 | 51,698 | 8,328 | 10,687 | 1,678 | 929,545 | 724 | 805,738 | 651 | 376,047 | 222 |
Finally, we addressed the false discovery rate in these analyses by generating a list of “impossible” junctions consisting of exon junction sequences joined in reverse order. For example, instead of splicing exon 1 to exon 3, we spliced exon 3 to exon 1. Using this approach, we found that 472 junctions were mapped to this set of 1,929,065 scrambled junctions, resulting in a false positive rate of 472/1,929,065 = 0.025%. This compares to 40,125 (or 2.08%) junctions that were mapped to the same number of “possible” junctions, indicating a false discovery rate of 472/40125 = 1.2% among mapped junctions.
Conclusions
High-throughput sequencing is able to generate sequence data equivalent to the entire EST collection, permitting both quantitative and structural analysis of the transcriptome in the cell. We have developed a molecular protocol, a computational pipeline for analysis of expression and alternative splicing, and a curve-fitting method for estimating the number of tags required for detecting specific genomic features, using the deep sequencing approach. On a prostate cancer cell model, our current analysis revealed a set of androgen-responsive genes. Our data also suggest prevalent poly(A)+ transcripts from both annotated protein-coding genes and intergenic regions and antisense transcripts, paving the way for further molecular analysis of regulated gene expression in mammalian cells. Curve-fitting analyses also reveal limitations in using deep sequencing to comprehensively detect alternative exons and splice junction sequences: Even with the tag density at which <1% additional discovery is achievable, we could only detect ≈20% of mRNA/EST-verified splice junctions compared with ≈70% of gene transcripts. Overcoming these limitations and performing comprehensive and quantitative measurements may require a combination of complementary tools like splice junction arrays that focus on annotated splice junctions (9, 26, 27).
Materials and Methods
Cell Culture, qPCR and Construction of cDNA Libraries.
Culturing LNCaP cells and DHT treatment were as described in ref. 28. Primers used for qPCR are listed in Table S4. Poly(A)+ RNA from LNCaP cells mock-treated and treated with DHT for 48 h was selected twice on oligo(dT) Dynabeads (Invitrogen). Purified poly(A)+ RNA (≈0.2 μg) was converted to cDNA with SuperScript III (Invitrogen) and 100 pmols of the first random primer (P1: 5′- AAT GAT ACG GCG ACC ACC GAN NNN NNN N-3′). cDNA was purified with the PCR purification kit (Qiagen), blocked at the 3′ end by terminal transferase reaction with ddNTPs, and immobilized on the streptavidin-coated magnetic beads (Invitrogen). cDNA on the beads were briefly washed 1× with 0.1 N NaOH and 2× with H2O before annealing of 100 pmols of the second random primer (P2: 5′-CAA GCA GAA GAC GGC ATA CGN NNN NNN N-3′) to the cDNA followed by extension with Taq polymerase at 25 °C for 30 min and 72 °C for 1 min. The beads were washed 2× with prewarmed wash buffer [10 mM Tris (pH 7.6), 1 mM EDTA, 0.1 M NaCl, 0.1% Tween 80] and the second strand DNA eluted with H2O at 95 °C for 10 min. The eluted DNA was PCR-amplified with AmpliTaq Gold (Perkin–Elmer), resolved on 2% agarose gel, eluted with the Qiagen PCR purification kit, and quantified on Nanodrop UV spectrometer. Approximately 0.4 fmol of PCR products was applied per lane on the Solexa flowcell for sequencing according to the manufacturer's instruction.
Genomic Mapping of the Sequence Tags.
Position-specific base compositions were made by compiling all uniquely aligned reads. The first base of every sequence tag was discarded because of nearly random utilization at the beginning of all sequences. To eliminate redundancies created by PCR amplification, all tags with identical sequences were considered single reads. After removal of adaptor sequences from the reads, the reads were compressed to a nonredundant list of unique sequence tags, which were then mapped to the human genome (hg17) with MosaikAligner (29), using a maximum of 2 mismatches over 95% alignment of the tag (34 nt) and a hash size of 15.
Transcriptome Analysis.
Genome sequences of human (hg17) and annotation for protein-coding genes were obtained from the University of California Santa Cruz (UCSC) (30). The lists of known human genes (knownGene containing 43,401 entries) and known isoforms (knownIsoforms containing 43,286 entries in 21,397 unique isoform clusters) with annotated exon alignments to human hg17 genomic sequence were processed as follows. Known genes that were mapped to different isoform clusters were discarded. All mRNAs aligned to hg17 that were >300 bases long were clustered together with the known isoforms. For the purposes of measuring differential gene expression, all genes were considered. For the purposes of inferring alternative splicing, genes containing <3 exons were removed from further consideration. A total of 2.7 million spliced ESTs were mapped onto the 17,478 high-quality gene clusters to identify alternative splicing. To eliminate redundancies in this analysis, final annotated gene regions were clustered together so that any overlapping portion of these databases was defined by a single genomic position.
An exon-body database (EBDB) was constructed as follows. Exons with canonical splice signals (GT-AG, AT-AC, GC-AG) were retained, resulting in a total of 213,736 exons. Of these, 92% of all exons were constitutive exons, 7% had evidence of exon-skipping, 1% exons were mutually exclusive alternative events, 3% exons had alternative 3′ splice sites, and 2% exons had alternative 5′ splice sites. An exon-junction database (EJDB) was constructed as follows. For each protein-coding gene, the 35 bases at the 3′ end of each exon were concatenated with the 35 bases at the 5′ end of the downstream exon. This was repeated, joining every exon of a gene to every exon downstream. This approach produced 1,929,065 theoretical splicing junctions. An equal number of “impossible” junctions was generated by joining the 35-base exon junction sequences in reverse order. MosaikAligner was used to align sequence tags to the junction database requiring no >2 mismatches over 95% of the sequence tag (34 nt). In addition, tags were required to have at least 4 nt across a specific splice junction. Junctions were annotated as constitutive (CE) if all of the exons within the span of the junction were annotated as constitutive (for a junction splicing exon 2 to 5, exons within the span are 2,3,4,5). Conversely, if any of the exons within the span of the junction were annotated as potentially spliced from EST or ACEscan annotations, the junction was labeled SE or ACE, respectively (Fig. S3).
To determine the number of tags contained within protein-coding genes, promoter, and intergenic regions, we arbitrarily defined promoter regions as 3 kb upstream of the transcriptional start site of the gene and intergenic regions as unannotated regions in the genome. Differentially expressed transcripts were identified by enumerating the number of tags that mapped within the spliced mRNA transcript in untreated and DHT-treated cells, using the total number of tags mapped to exons in each condition as a basis for determining significance by the χ2 statistic. Comparison of differentially expressed genes (P < 0.01) to published datasets of hormone-regulated genes was performed as follows. For each of 1,000 iterations, an equal number of genes to our list of differentially expressed genes were randomly selected and compared with a published list of hormone-regulated genes. The number overlapped was recorded, and the mean and standard deviation of 1000 iterations was used to compute a Z score, resulting in a P value computed assuming a standard normal distribution. GO analysis was performed as described in ref. 25.
Curve-Fitting Method to Estimate Saturating Tag Density and Observable Features.
Tags were randomly sampled into subsets representing 10%, 20% etc. of the total number of sequence tags available. These were aligned as described above and the number of features detected was assessed. To determine the number of sequence tags required to reach a user-defined threshold for saturation, the percentage change in discovering additional features was determined as follows:
where T(n) is the number of tags, s is the sampling size (in our case, 2 million tags), n is a constant multiplier, C(n) is the empirical change in number of features detected, and F(n) is the number of empirical features detected at n. A scatter plot of C(n) to T(n) was fitted with a power curve of the form c(n) = a × T(n)b and an exponential curve of the form c(n) = aebT(n), where c(n) is the change estimated by the curve fitting.
The equation that had the best fit, indicated by R2, was used to extrapolate the tag density required to achieve a defined change in the number of features detected. The number of estimated features was calculated by
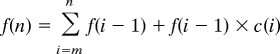 |
where m is user-defined (in our case, m = 6).
Supplementary Material
Acknowledgments.
We thank F.H.G. for his generous support and encouragement during the course of this investigation when G.W.Y. was a Crick–Jacobs Junior Fellow at the Salk Institute. M.G.R. is an Investigator of the Howard Hughes Medical Institute. This work was supported by a Prostate Cancer Foundation award (to M.G.R. and X.D.F.) and National Institutes of Health Grants GM052872 (to X.D.F.) and HG004659 (to G.W.Y. and X.-D.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807121105/DCSupplemental.
References
- 1.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 2.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 5.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 6.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 7.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 10.Imanishi T, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 13.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perocchi F, Xu Z, Clauder-Munster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 16.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 18.Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. Transcriptional interference: An unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 19.Bainbridge MN, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velasco AM, et al. Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinology. 2004;145:3913–3924. doi: 10.1210/en.2004-0311. [DOI] [PubMed] [Google Scholar]
- 21.Segawa T, et al. Androgen-induced expression of endoplasmic reticulum (ER) stress response genes in prostate cancer cells. Oncogene. 2002;21:8749–8758. doi: 10.1038/sj.onc.1205992. [DOI] [PubMed] [Google Scholar]
- 22.Nelson PS, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci USA. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePrimo SE, et al. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3:RESEARCH0032. doi: 10.1186/gb-2002-3-7-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, et al. Quantitative expression profile of androgen-regulated genes in prostate cancer cells and identification of prostate-specific genes. Int J Cancer. 2001;92:322–328. doi: 10.1002/ijc.1196. [DOI] [PubMed] [Google Scholar]
- 25.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 27.Pan Q, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Li HR, et al. Two-dimensional transcriptome profiling: Identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66:4079–4088. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]
- 29.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 30.Karolchik D, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.