Abstract
Brain structure and function experience dramatic changes from embryonic to postnatal development. Microarray analyses have detected differential gene expression at different stages and in disease models, but gene expression information during early brain development is limited. We have generated >27 million reads to identify mRNAs from the mouse cortex for >16,000 genes at either embryonic day 18 (E18) or postnatal day 7 (P7), a period of significant synaptogenesis for neural circuit formation. In addition, we devised strategies to detect alternative splice forms and uncovered more splice variants. We observed differential expression of 3,758 genes between the 2 stages, many with known functions or predicted to be important for neural development. Neurogenesis-related genes, such as those encoding Sox4, Sox11, and zinc-finger proteins, were more highly expressed at E18 than at P7. In contrast, the genes encoding synaptic proteins such as synaptotagmin, complexin 2, and syntaxin were up-regulated from E18 to P7. We also found that several neurological disorder-related genes were highly expressed at E18. Our transcriptome analysis may serve as a blueprint for gene expression pattern and provide functional clues of previously unknown genes and disease-related genes during early brain development.
Keywords: E18, P7, brain, transcription factors, neural diseases
Mammalian brain development can be largely divided into 2 periods: embryonic and postnatal. Embryonic mouse brain development starts ≈10–11 days after gestation (E10–E11) with massive neuronal production from neural stem cells. The development of rodent cerebral cortex is a well-studied model system, where the initial neurons form the subplate layer, although subsequent neurons migrate in an inside-out pattern to form the multilayer cortical structure (1, 2). By embryonic day 18 (E18), neurons start to send out axons and dendrites to be poised for synaptic connections. After birth, the first week of postnatal brain development is characterized by elevated production of astrocytes, which are crucial for neuronal synaptogenesis (3, 4). By postnatal day 7, many neurons start to establish synaptic connections with other neurons, forming a primitive neural circuit.
Early brain development is precisely controlled by transcription factors, cell adhesion molecules, receptors and channels, synaptic proteins, and other effectors. A single misstep might result in a severe deformation of the brain circuit. For example, loss of Otx2 function results in the absence of early brain development (5). Since many psychiatric disorders such as autism and mental retardation are closely associated with early brain development, understanding the gene expression profile will facilitate the search for an optimal treatment for these disorders. Previous surveys of early brain development have focused on a small number of genes. More recently, microarray studies and others have revealed differential expression of groups of genes in specific brain regions or associated with brain disorders (6–14). Specifically, gene expression has been examined using whole brains or brain tissues from young and old, and diseased mice (7, 8, 12, 15). In a microarray study of 11,000 genes and ESTs, the expression level of 1,926 genes was found to change significantly during hippocampal development from E16 through P30 (11). Also, 366 ProbeSets were found to be differentially expressed during postnatal 2–10 weeks (13). However, microarray technology has several limitations: (1) the number of genes is fixed; (2) the sensitivity is limited by background hybridization; and (3) the inaccuracy of mRNA levels because of difference in hybridization among probes.
Recent technologies have allowed massive amounts of sequencing at relatively low cost (16–18) and analysis of gene expression by sequencing is highly reproducible and more sensitive than microarrays (19). In particular, the Solexa/Illumina sequencing technology has advantages in high coverage and relatively low cost (16–18). It has been used to interrogate transcriptomes of yeast, mouse, and human tissues (20–23). However, previous studies have not analyzed the change of transcriptome during early brain development. Because the change of expression level could provide clues to gene function, large-scale sequencing to detect gene expression has great potential in finding important genes for development. We have conducted Solexa/Illumina sequencing of cDNAs from E18 and P7 mouse brain cortices, and report here the detection of >16,000 genes, with 3,758 being differentially expressed between these 2 stages. In addition, we report on the discovery of previously unknown splice variants. Our results pave the way for further functional analysis of a large number of genes in the early developing brain.
Results and Discussion
Isolation of RNAs from Dissected Mouse Brain Cortex Tissues and Library Construction.
We compared transcriptomes of 2 important stages of developing mouse brain, E18 and P7, characterized by the production of the majority of neuronal cells and significant synaptic contacts between neurons, respectively. For each of 2 biological replicates, cortices were dissected from 5–8 E18 embryos and P7 pups, and used to isolate total RNAs and mRNAs, which were used to generate cDNA libraries without vectors. A small portion of each library was cloned into a plasmid vector and several clones were sequenced to determine the quality of the cDNAs. The cDNA libraries were then used for high-throughput sequencing.
High-Throughput Sequencing and Mapping of the Reads.
High-throughput sequencing of the cDNA libraries were carried out using the Solexa/Illumina technology. Because this technology was still in its early stages of application, the initial sequencing was done using single-end cDNA libraries from 1 biological replicate of mouse brain at the E18 and P7 stages. After analysis of the single-end cDNAs, cDNAs from the same 2 mRNA samples were used to construct libraries for paired-end sequencing. From these experiments, the single-end sequencing produced 2,956,444 reads from the E18 and 3,619,970 from P7 stages, respectively. The paired-end round generated 4,536,964 reads from the E18 library and 4,019,273 reads from the P7 library. A second biological replicate of mouse brain cortices at the E18 and P7 stages was subjected to paired-end sequencing, resulting in 5,501,311 reads at E18 and 6,042,658 reads at P7 (Table 1). For both single and paired-end sequences we examined the quality of the base calls using Galaxy (http://galaxy.psu.edu) (24, 25). The distribution of quality score [Phred-equivalent metric (26, 27)] at each base of the read (Fig. S1) indicated that bases before 28 generally had quality scores of 20 or higher; therefore, only the first 27 high-quality bps were used in the subsequent analysis to maximize the sequence reliability, while retaining sufficient length for analyses.
Table 1.
Summary of read number
| Reads category | Biological replicate 1 |
Biological replicate 2 |
||||
|---|---|---|---|---|---|---|
| Single-end |
Paired-end* |
Paired-end |
||||
| E18 | P7 | E18 | P7 | E18 | P7 | |
| Total reads | 2,956,444 | 3,619,970 | 4,536,964 | 4,019,273 | 5,501,311 | 6,402,658 |
| Uniquely mapped | 1,886,668 | 2,117,328 | 2,329,560 | 2,238,480 | 2,784,748 | 3,220,724 |
| In exons | 1,099,869 | 1,404,798 | 1,376,421 | 1,577,198 | 1,948,700 | 2,448,499 |
| In novel transcripts | 2,302 | 2,689 | 621 | 746 | 794 | 1,084 |
*For paired-end data, 1 pair of sequencing results was counted as 1 read. Only if both ends were uniquely mapped, we count this pair as the uniquely mapped read.
To identify genes expression in the mouse brain cortex, we mapped the reads against the July 2007 assembly of the mouse genome using the RMAP (28) tool specifically designed for Illumina (Solexa) data. As shown in Table 1, 60.9% of single-end reads were uniquely mapped and there were 51.8% of paired-end reads that were mapped uniquely at both ends.
We compared the genomic coordinates of reads against the gene locations of UCSC (University of California at Santa Cruz) Known Gene collection (29, 30) (Referred to as “Known Genes” in the remainder of this study). Because the UCSC Genes dataset is highly redundant because of multiple splice variants, we aggregated the reads for each gene cluster—a collection of transcripts representing a single gene. Interestingly, besides the large number of reads that mapped to annotated exons, additional large number of reads mapped to introns and intergenic regions (Table 1), consistent with the previous discovery of pervasive transcription of the human genome (31). Finally the number of reads matching each gene was calculated for all of the Known Genes. An example of how reads were related to exons and genes is shown in Fig. 1. The per-gene distribution of reads was highly consistent between the 2 paired-end data, with Pearson's correlation coefficient being 0.97 and 0.96 for E18 and P7, respectively (Fig. 2 A and B). The single-end data and paired-end data were also highly consistent in which Pearson's correlation coefficient was 0.96 and 0.95 for E18 and P7 stages, indicating that both the single-end and paired-end sequencing yielded similar results (Fig. S2A and B). This consistency was in agreement with the high reproducibility reported recently (19–23) and permitted the detection of differential expression and analysis of alternative transcripts. Interestingly, the distributions of the number of genes according to read counts were very similar between the 2 replicates for each stage, and even between the 2 stages (Fig. 2C).
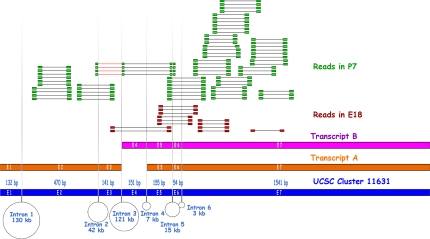
Fig. 1.
An example of mapping reads to a gene. The UCSC gene/cluster 11631 has 2 transcripts, A and B. Transcript A has exons 1, 2, 3, 5, 6, and 7. Transcript B has exons 4, 5, 6, and 7. Thirteen paired-end reads (red) mapped to this gene at E18 and 65 paired-end reads (green) mapped to this gene at P7. At P7 stage, some reads mapped to transcript-specific exons 1 and 2, providing evidence for the expression of transcript A. Four reads mapped to an unknown junction (the red line) between exons 2 and 4, indicating a possibly unreported transcript at P7 stage. Because the cDNA synthesis was polyT primed, there were more reads in 3′ region than 5′.
Fig. 2.
A comparison between 2 biological replicates and among all datasets. (A) The comparison of reads per gene between the first and second paired-end data in E18. Since the read number per gene ranges from 0 to >10,000, the read numbers adding 1 were transformed by log10. There is a good correlation between the first and second paired (R = 0.97). (B) The comparison of reads per gene between the first and second paired-end data in P7 (R = 0.96). (C) The line chart showing the distribution of genes with different reads. The y axis is the number of genes. The x axis is different intervals of read number. The number of genes at E18 and P7 showed parallel changes in both single-end and paired-end analysis. Each biological replicate at each stage contained the empbryos of pups from a single female mouse; these embryos/pups were probably more similar than embryos/pups from different litters. It is possible that potential maternal effects, such as age and other variability, might contribute to the results, which nevertheless are highly reproducible between 2 different mothers for each stage.
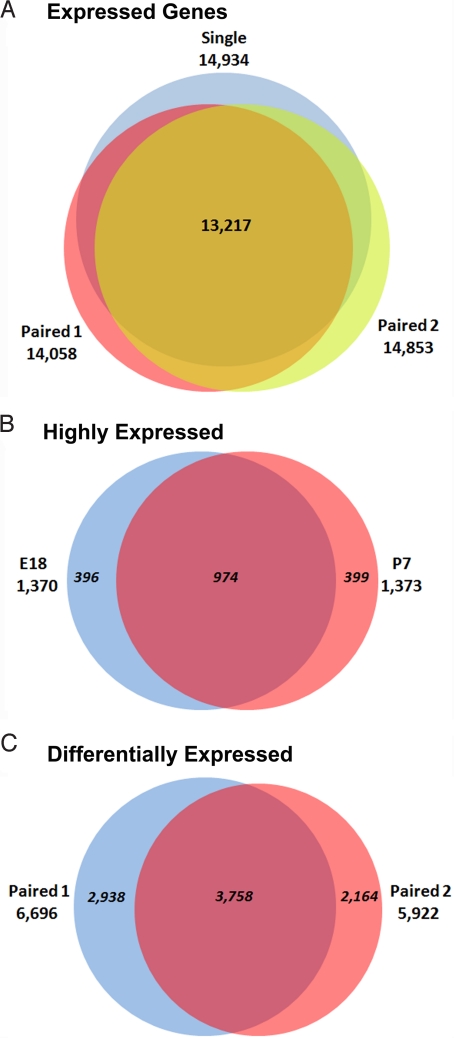
To minimize false positives for expressed genes, we required at least 2 uniquely mapped reads as detectable expression of a given gene. The single-end and paired-end sequences yielded similar numbers of detected genes (Fig. S2 C and D). Our single-end reads revealed the expression for 13,463 and 14,243 genes at E18 and P7, respectively. From the first paired-end analysis, we identified reads for 12,590 genes from the E18 cortex and 12,991 genes from P7. From the second paired-end dataset, 13,642 and 14,223 genes showed expression at E18 and P7, respectively. Altogether, we detected the expression of 14,787 genes at E18 and 15,423 genes at P7, with a total of 16,083 genes, representing 58.7% of mouse Known Genes (Fig. 3A). In addition to the large overlap between the number of expressed genes in E18 and P7, there were also a substantial number of genes that are preferentially expressed at a particular stage (660 at E18; 1,296 at P7). If we regarded the top 5% genes among total clusters of UCSC Known Genes (27, 389) as highly expressed, we found that besides a large number of genes (974) expressed at both stages, an appreciable number of highly expressed genes were also uniquely expressed at each stage (396 at E18; 399 at P7) (Fig. 3B and Fig. S2E).
Fig. 3.
Venn diagrams showing the number of expressed genes. (A) The number of expressed genes in 3 analyses, with a total of 16,083. (B) The number of highly expressed genes. The top 5% genes were regarded as highly expressed ones. The 2 paired-end data were summed. (C) The number of differentially expressed genes between E18 and P7. There were a substantial number of differentially expressed genes (3, 758) supported by both paired-end analyses.
Strong Evidence for Differential Gene Expression.
The large numbers of sequencing reads represent a deep sampling of the transcriptome and can be an excellent digital measure of the relative abundance of transcripts (20). To obtain statistical support for the differences between the 2 stages, we applied Fisher's exact test on the read count of each gene at E18 and P7. We performed the test in 2 different ways to obtain relatively conservative estimates. To balance false positive and false negative results, we summed reads from our 2 paired-end biological replicates and then performed the test. A large number (7,688) of genes were found to be differentially expressed between E18 and P7 (at 0.01 significance level). In a more stringent analysis, we applied the test on each paired-end dataset separately and then found the intersection between the 2 replicates that was significant and concordant in the direction of differential expression in both replicates. There were still 3,758 differentially expressed genes with significance level of 0.01 or less in both replicates (Fig. 3C and Table S1). We also performed an analysis that combined the data from both biological replicates using LIMMA (see Materials and Methods). Although LIMMA is less powerful than Fisher's exact test in detecting differential expression, LIMMA allows a combined analysis of the replicates. Our LIMMA analysis showed that 5,811 genes had a q value between 0.10 and 0.15, a moderate support for differential expression. These 5,811 genes included 3,337 (88.8%) of the above-mentioned 3,758 differentially expressed genes from the Fisher's exact test with data of both replicates. Therefore, these 2 types of analyses gave generally consistent results, and represent statistically well-supported findings of differential gene expression. Many of the differentially expressed genes were known to play important roles in neuronal network formation. Strikingly, approximately 500 genes showed dramatic changes in expression level but did not have functional annotation in the UCSC Known Genes.
Differentially Expressed and Unreported Splice Variants.
Many mammalian genes are known to have alternatively spliced transcripts, which are usually not specifically detected in microarray experiments. Specific splice variant(s) can be detected by the reads that map to exon(s) or exon combination(s) that are unique to a transcript. Such exonic sequences are markers allowing the comparison of splice variants between 2 samples (Fig. 1). In addition to reads mapped to unique exon-exon junctions, the paired-end reads can identify splicing variants if the 2 ends map to 2 exons in a transcript-specific combination. Our single-end reads allowed the detection of alternatively spliced transcript(s) for 4,131 and 4,169 genes at E18 and P7, respectively. Furthermore, alternative spliced transcripts for 2,112 genes were detected in only 1 of these stages. Similarly, paired-end reads of the first replicate supported alternatively spliced transcript(s) for 4,062 and 4032 genes at E18 and P7, respectively. Again, at least 1 alternatively spliced transcript for each of 2,557 genes was detected in only 1 of the 2 stages. The second paired-end replicate uncovered alternative splicing of 4,354 and 4,496 genes, respectively, at E18 and P7, among which 2,055 genes had alternatively spliced transcript(s) detected in only 1 of the stages. Among the several thousand genes detected to have alternative splice variants, 320 genes had splicing differences that were supported by all 3 sequencing datasets. A recent study also employing Solexa sequencing found that 92%–94% of human genes had alternative splicing among 15 types of tissues (32). Our results show that there is also substantial splicing variation between 2 developmental stages of the same tissue.
To detect unknown exon-exon junctions, we searched for reads that mapped on alternative junctions from known ones. In addition, a pair of ends that mapped in 2 exons could support unreported junctions if they did not belong to the same known transcripts (Fig. S3). We modified a previous procedure (20) to incorporate both single-end and paired-end reads (see Materials and Methods). Single- and paired-end junction analyses differed in 3 ways: (1) the target pseudosequences were different; (2) paired-end data allowed the detection of unknown splicing forms when both ends mapped 2 exons in a reported combination; (3) paired-end analysis required both ends mapping onto the same transcript. In single-end analysis, we found that 2,302 and 2,689 reads mapped to junctions distinct from known ones in 1,367 and 1,596 genes at E18 and P7, respectively. In the first paired-end analysis, we found 621 reads as evidence of unreported transcripts in 143 genes at E18. At P7, we found 746 reads supporting unreported transcripts in 172 genes. From the second paired-end analysis, we identified 794 and 1084 reads for unknown transcripts in 448 and 536 genes at E18 and P7, respectively (Table 1 and Table S2). In total, we identified unreported splicing forms in 2,930 genes from the single end and 2 paired-end samples, with 974 genes supported by 2 or more datasets (Table S3). Among them were genes that play important roles in the brain, such as FBXO2 (maintaining postmitotic neurons), App (amyloid beta (A4) precursor protein, involved in plasticity and Alzheimer's), Aplp1 (synaptic maturation), and BPTF (gene regulation, related to Alzheimer's disease).
The Most Expressed Genes During Early Brain Development.
Our analysis showed that genes with highest early brain expression level were responsible for ATP production in mitochondria or encoding microtubule proteins. Among the top 10 most expressed genes at E18 and P7 from single-end analysis (most reads), 7 genes were the same: NADH dehydrogenase 1, cytochrome b, NADH dehydrogenase 4, NADH-ubiquinone oxidoreductase chain 2, cytochrome c oxidase subunit I, tubulin β5, and microtubule-associated protein tau. Paired-end analysis also showed that these genes were among most expressed ones except for minor changes in the ranking. The results are consistent with the need for mitochondrial biogenesis and energy during active cell proliferation and growth in early brain development. Microtubules are crucial both for cell division and differentiation generally and for neuronal axon and dendritic growth specifically. At P7, an unclassified gene AK157178 in mouse (ortholog of human WASF2/SCAR2/WAVE2) is also among the top 10 expressed genes, suggesting an important function during early brain development and warrants further studies.
Developmental Regulation of Genes Encoding Synaptic Proteins and Receptors.
Consistent with a significant increase in synaptogenesis during neonatal brain development, we found that many synaptic protein genes and receptor genes were substantially up-regulated from E18 to P7 (Table S4). Genes for synaptic proteins were already expressed at modest to high level at E18 and were further increased at P7. The highest among these at P7 encoded synaptophysin, complexin 2, syntaxin 1A, and synucleins (paired-end analysis). The observation that many genes coding for synaptic proteins were almost uniformly up-regulated by 4- to 5-fold from E18 to P7 suggests a potentially common mechanism for transcriptional regulation. Similarly, genes for neurotransmitter receptors were also up-regulated in general, but different subunits showed some distinct developmental changes (Table S5). Among genes encoding glutamate receptors, which mediate the majority of neuronal excitation, GluR2 for the AMPA receptor subunit 2 was highly expressed, whereas GluR1, 3, and 4 were modestly expressed at E18. Interestingly, from E18 to P7, GluR2 was down-regulated but GluR1 was up-regulated. Among genes for NMDA receptor subunits, NR1A and NR2B were significantly expressed and up-regulated from E18 to P7, but NR2A, NR2C, and NR2D had very low levels at both stages. NR2A expression will likely be up-regulated during later brain development (33). Among metabotropic glutamate receptor genes, mGluR5 was most highly expressed at E18 and further increased at P7. GABA receptors mediate the major inhibition in the brain. Interestingly, genes for GABAA receptor α subunits were expressed at low or modest (α5) level in E18 and P7, whereas those for GABAB receptors were highly expressed at E18 and further increased at P7, suggesting an important role in the early brain. In contrast, most genes encoding receptors for glycine, acetylcholine, dopamine, and serotonin were detected at low levels (Table S5).
Among voltage-dependent channels, genes for the majority of Na+, K+, and Ca2+ channels were expressed at low levels in E18 but generally up-regulated by P7 (Table S5). For voltage-gated Ca2+ channels, genes for low-voltage sensitive T-type channel α subunits were expressed more than those for L-, N-, and P/Q-type channels at E18, but the latter ones were significantly up-regulated from E18 to P7. For K+ channels, Kcnab2, Kcnc4, Kcnd2, Kcnf1, Kcnh3, Kcnj4, and Kcnq2 were the most highly expressed genes. For Na+ channels, Scn2α1, Scn8α, Scn1β, Scn2β, and Scn3β were all highly expressed, and up-regulated from E18 to P7 (Table S5).
Expression of Cell Signaling Genes.
Among cell signaling molecules, calmodulin 3, adenylate cyclase 1, and calmodulin kinase-like vesicle-associated protein were encoded by the top expressing genes that were highly up-regulated from E18 to P7 (Table S4). Other genes encoding protein kinases, such as PKC β1 and γ, CaMKII, and protein tyrosine kinase 2β and 3, together with those for protein phosphatase 1 and protein tyrosine phosphatase N, were also up-regulated by >4-fold from E18 to P7.
Wnt, BMP (bone morphogenetic protein), and hedgehog are known to be involved in early brain development (36, 37). Among all Wnts detected, only Wnt7b was significantly expressed and Wnt7a modestly expressed at E18, and both slightly down-regulated at P7 (Table S5). Wnt4 was up-regulated from E18 to P7, at modest levels. For the BMP group, only BMP1 was modestly expressed at similar levels in the E18 and P7 mouse cortices. Consistently, we found that only BMP1a and 1b receptors were modestly expressed. Other BMPs were generally at very low expression level. Similarly, the sonic hedgehog gene, which is crucial for early embryogenesis, was substantially reduced to almost undetectable level at P7 (Table S5).
Detecting a Large Number of Genes Encoding Transcription Factors.
Transcription factors (TFs) perform important regulatory functions by controlling a variety of cellular processes. In the mouse genome, 1,445 genes were identified to encode TFs and 983 were expressed in the brain (34). Our sequence data uncovered expression of at least 1,024 genes encoding TFs at the E18 stage and 1039 genes at the P7 stage, totaling 1,079 genes. Among these, 559 and 504 genes were detected with at least 50 reads from combined single-end and paired-end datasets at E18 and P7, respectively. Interestingly, many of the most highly expressed TF genes at both E18 and P7 stages are the same, including Thra, Tcf4, Pbx1, Nr2f1, Sox11, Sox4, and Bcl11b. Among the 349 differentially expressed TF genes supported by both replicates, a large majority showed higher levels at E18 than P7, suggesting that transcriptional regulation is important for active neuronal cell division and differentiation, more so than the later neuronal maturation stage (Table S5). Many of the genes that showed highest ratios of read numbers at E18 to P7 encoded zinc finger proteins; among 117 differentially expressed zinc-finger genes, 56 were more highly expressed at E18, suggesting that they play crucial roles in embryonic neuronal development. In addition, the expression of Otx2, encoding a TF required for initial forebrain development (5), was substantially reduced at P7 (Table S5). A recent report found that Otx2 is expressed again at relatively high levels later in the visual cortex during the critical period of P28-P30 (35).
Detection of Genes for Autophagy and Apoptosis.
Although autophagy has been reported to participate in many biological processes, its exact role in vertebrate development, especially neurodevelopment, is far from fully characterized (38). A recent study found that Ambra1 regulates autophagy and is critical for neural tube development (39). Our result showed that Ambra1 has appreciable expression levels in both E18 and P7. Besides Ambra1, there are 17 other autophagy-related genes (Table S5). Nine of these had similar or higher expression levels than Ambra1. Especially, Atg9a showed significant increase in expression level from E18 to P7. Apoptosis is important for development and is also associated with neurodegenerative diseases (40). Among genes related to apoptosis (Table S5), the gene for Cugbp2/Napor-1, an apoptosis related RNA-binding protein, had extraordinarily high expression levels at both E18 and P7. In addition, the Bcl family genes Bcl11a, Bcl11b, and Bcl7a were highly expressed. These highly expressed apoptosis-related genes were often down-regulated from E18 to P7, suggesting that programmed cell death is more active at E18 than P7. In addition, many other genes annotated to participate in apoptosis also had substantial expression level Table S5), suggesting the important role of apoptosis in embryonic neuronal development.
Up- and Down-regulated Genes and Previously Unknown Genes.
Many transporter genes showed greatly increased expression from E18 to P7 (Table S4). For example, Na+/K+-ATPases were most abundantly expressed at E18 already and further increased by 4–5-fold at P7, consistent with their critical functions in maintenance of resting membrane potential and cell volume. We have also identified a group of genes that are substantially down-regulated from E18 to P7 (Table S4). As noted above, 2 genes encoding the transcription factors Sox4 and Sox11 were among the mostly highly expressed transcription factor genes at both stages. These genes are known to function in cell fate determination and were more highly expressed at E18 than at P7 (6–10-fold down-regulation), supporting their greater roles in early neural differentiation than neuronal maturation. A group of genes with unknown functions were also identified that display substantial up- or down-regulation from E18 to P7 (Table S4).
Genes Related to Neurological Disorders.
Among the highly expressed genes during early brain development, we further identified genes associated with neurological diseases (Table S5). The APP gene (amyloid beta precursor protein) was already highly expressed at E18, and further up-regulated at P7. Cleavage of APP by β and γ secretases results in amyloid β peptide deposits in senile brains, especially among Alzheimer's patients, but the high expression of APP in E18 and P7 cortex suggests an uncharacterized function in brain development. Some mental retardation (Atrx) and seizure-related genes (Sez6) were highly expressed from E18 to P7, suggesting a possible link to infant brain defects or infant seizures if these genes do not function properly. Interestingly, autism related gene (Auts2) was highly expressed at E18 but substantially down-regulated by P7, raising the possibility that continuous expression of this gene might be associated with autism. Some prion protein genes (Prnp and Prnpip1) were also highly expressed at E18 and further up-regulated by P7, suggesting a potential function of these genes in neuronal maturation or synapse formation and plasticity.
Conclusion
Our high-throughput sequence analysis of the transcriptome of the mouse developing cortices detected the expression of >16,000 genes and uncovered 3,758 genes that were differentially expressed between the E18 and P7 stages. The methods we used and the comparison between single-end and paired-end analyses will provide foundation for further development of bioinformatic tools to analyze next-generation sequencing data. The expression information suggested important functions for a number of regulatory genes, and provided strong evidence for a highly dynamic transcriptome during mouse brain development. Previous microarray work in postnatal cortical development found an increase of sox11 but decrease of sox4 expression from postnatal week 2 to week 3 (13). However, our results revealed a substantial decrease of both sox11 (10-fold) and sox4 (5-fold) from E18 to P7, suggesting that transcriptional regulation may differ significantly before and after birth. The detection by our deep sequencing data of the expression of a majority of the known transcription factor genes during E18–P7 stages further suggested that the transcriptional regulation plays a prominent role during a critical window of brain circuit formation.
Materials and Methods
Mouse Brain Dissection, RNA Extraction, cDNA Synthesis, and Sequencing.
The animal protocol was approved by the Penn State University IACUC committee. A total of 4 pregnant female mice (C57BL/6J, 10–12 weeks old), purchased at 2 different time points as biological replicates, were from Charles River Laboratories at 14-days pregnancy. The mice were fed with Purina laboratory rodent diet 5001. For each replicate of 2 female mice, 1 was killed with CO2 at 18-days pregnancy to collect the E18 embryos, and the other was allowed to give birth to collect pups at P7. Six to eight of the E18 embryos and 5 to 6 P7 pups were decapitated, and cortical hemispheres were collected by removing the brainstem, cerebellum, and midbrain. The cortices of all embryos (or all pups) from each litter were dissected, immediately frozen in liquid nitrogen, and stored at −80 °C before RNA extraction. Total RNA was isolated from approximately 260 and 750 mg of E18 and P7 cortical tissues, respectively, using an RNA isolation kit from Ambion Inc. according to manufacturer's protocol, yielding 1.2 and 5.4 mg of RNA, respectively. The RNA samples were treated with DNase I (Invitrogen), then sent to Fasteris SA for mRNA purification and cDNA library construction for sequencing using the Illumina/Solexa technology.
Sequencing Quality, Reads Mapping, and Sequence Analyses.
The quality of sequencing result was summarized and plotted using Galaxy (http://galaxy.psu.edu). The mouse genome sequence of July 2007 assembly was downloaded from UCSC genome informatics portal (http://genome.ucsc.edu/). RMAP (http://rulai.cshl.edu/rmap/) reports the uniquely mappable and ambiguous reads into 2 separate files. To simplify subsequent analysis, the chromosome sequences were concatenated into 1 pseudosequence by self-developed scripts (these and others are available upon request) and then single-end and paired-end reads were mapped onto the concatenated sequence. We only used the uniquely mapped reads in the subsequent analysis. For paired-end reads, we required both ends to be uniquely mapped.
The exon coordinates were downloaded from the UCSC table browser and analyzed using scripts developed here. The number of reads that matched each exon and UCSC transcript cluster was calculated. Fisher's exact test was applied with R (http://www.r-project.org/) and “sagenhaft” library (http://tagcalling.mbgproject.org) in Bioconductor to identify statistically significant differentially expressed clusters between P7 and E18. Also, for sufficiently highly expressed genes, read counts were converted to percentage of total mapped reads, and a supplementary analysis of the log2 (percentages) was done as a randomized complete block design using the “LIMMA” library (41) in Bioconductor. The reads mapped to transcript-specific sequence were selected (see SI Methods for the selection criteria). Using read mapping information, transcripts were compared for splicing events between P7 and E18. In single-end analysis, 20 bp on either side of all possible junctions between known exons were connected into 1 pseudosequence. In paired-end analysis, the 20 bp flanking possible junctions and the full-length exonic sequences were connected into 1 pseudosequence (see SI Methods). We then mapped our reads to this concatenated junction sequence using RMAP, separately from the genomic mapping. Reads that were already mapped in the genomic analyses were excluded.
Supplementary Material
Acknowledgments.
We thank Yi Hu for technical assistance and the anonymous reviewers for helpful comments on a previous version of the manuscript. This work was supported by the Biology Department, Eberly College of Sciences, and Huck Institute of the Life Sciences, Pennsylvania State University (H.M.); National Institutes of Health Grant NS054858; and National Science Foundation Grants 0821955 (to G.C.) and DBI-0543285 (to A.N.). X.H. was supported by the Intercollege Graduate Program in Genetics and the Biology Department, Pennsylvania State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902417106/DCSupplemental.
References
- 1.Allendoerfer KL, Shatz CJ. The subplate, a transient neocortical structure: Its role in the development of connections between thalamus and cortex. Annu Rev Neurosci. 1994;17:185–218. doi: 10.1146/annurev.ne.17.030194.001153. [DOI] [PubMed] [Google Scholar]
- 2.Rakic P. A century of progress in corticoneurogenesis: From silver impregnation to genetic engineering. Cereb Cortex. 2006;16(Suppl 1):i3–17. doi: 10.1093/cercor/bhk036. [DOI] [PubMed] [Google Scholar]
- 3.Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Ullian EM, Christopherson KS, Barres BA. Role for glia in synaptogenesis. Glia. 2004;47:209–216. doi: 10.1002/glia.20082. [DOI] [PubMed] [Google Scholar]
- 5.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 6.Funatsu N, Inoue T, Nakamura S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: Identification of several region- and layer-specific genes. Cereb Cortex. 2004;14:1031–1044. doi: 10.1093/cercor/bhh063. [DOI] [PubMed] [Google Scholar]
- 7.Jiang CH, Tsien JZ, Schultz PG, Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci USA. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the aging brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 9.Miki R, et al. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc Natl Acad Sci USA. 2001;98:2199–2204. doi: 10.1073/pnas.041605498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirnics ZK, et al. DNA microarray profiling of developing PS1-deficient mouse brain reveals complex and coregulated expression changes. Mol Psychiatry. 2003;8:863–878. doi: 10.1038/sj.mp.4001389. [DOI] [PubMed] [Google Scholar]
- 11.Mody M, et al. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001;2:RESEARCH0042. doi: 10.1186/gb-2001-2-10-research0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semeralul MO, et al. Microarray analysis of the developing cortex. J Neurobiol. 2006;66:1646–1658. doi: 10.1002/neu.20302. [DOI] [PubMed] [Google Scholar]
- 14.Zapala MA, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci USA. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18:839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
- 17.von Bubnoff A. Next-generation sequencing: The race is on. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Strausberg RL, Levy S, Rogers YH. Emerging DNA sequencing technologies for human genomic medicine. Drug Discov Today. 2008;13:569–577. doi: 10.1016/j.drudis.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 23.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 24.Blankenberg D, et al. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giardine B, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 27.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu F, et al. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 30.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: Diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 34.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 35.Sugiyama S, et al. Experience-dependent transfer of Otx2 homeoprotein into the visual cortex activates postnatal plasticity. Cell. 2008;134:508–520. doi: 10.1016/j.cell.2008.05.054. [DOI] [PubMed] [Google Scholar]
- 36.Salinas PC, Zou Y. Wnt signaling in neural circuit assembly. Annu Rev Neurosci. 2008;31:339–358. doi: 10.1146/annurev.neuro.31.060407.125649. [DOI] [PubMed] [Google Scholar]
- 37.Charron F, Tessier-Lavigne M. Novel brain wiring functions for classical morphogens: A role as graded positional cues in axon guidance. Development. 2005;132:2251–2262. doi: 10.1242/dev.01830. [DOI] [PubMed] [Google Scholar]
- 38.Cecconi F, et al. A novel role for autophagy in neurodevelopment. Autophagy. 2007;3:506–508. doi: 10.4161/auto.4616. [DOI] [PubMed] [Google Scholar]
- 39.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 40.Yeo W, Gautier J. Early neural cell death: Dying to become neurons. Dev Biol. 2004;274:233–244. doi: 10.1016/j.ydbio.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistl Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.