Abstract
The success of prenatal carrier screening as a disease prevention strategy in the Ashkenazi Jewish (AJ) population has driven the expansion of screening panels as disease-causing founder mutations have been identified. However, the carrier frequencies of many of these mutations have not been reported in large AJ cohorts. We determined the carrier frequencies of over 100 mutations for 16 recessive disorders in the New York metropolitan area AJ population. Among the 100% AJ-descended individuals, screening for 16 disorders resulted in ~1 in 3.3 being a carrier for one disease and ~1 in 24 for two diseases. The carrier frequencies ranged from 0.066 (1 in 15.2; Gaucher disease) to 0.006 (1 in 168; nemaline myopathy), which averaged ~15% higher than those for all screenees. Importantly, over 95% of screenees chose to be screened for all possible AJ diseases, including disorders with lower carrier frequencies and/or detectability. Carrier screening also identified rare individuals homozygous for disease-causing mutations who had previously unrecognized clinical manifestations. Additionally, prenatal testing results and experience for all 16 disorders (n = 574) are reported. Together, these data indicate the general acceptance, carrier frequencies, and prenatal testing results for an expanded panel of 16 diseases in the AJ population.
Keywords: Ashkenazi Jewish, carrier screening, carrier frequency, residual risk, prenatal diagnosis
INTRODUCTION
Prenatal carrier screening for severely debilitating recessive genetic diseases prevalent in the Ashkenazi Jewish (AJ) population began with Tay-Sachs disease (TSD; MIM# 272800) in the early 1970s [Kaback et al., 1993; Kaback, 2000]. Since that time the number of disorders included in AJ screening panels increased as the causative genes and founder mutations for additional diseases made DNA-based carrier testing feasible. Coupled with genetic counseling, prenatal carrier screening has been widely accepted by the AJ community [Eng and Desnick, 2001; Eng et al., 1997], and in 2004, the American College of Obstetricians and Gynecologists (ACOG) updated its recommended AJ carrier screening panel to include four highly prevalent neurodegenerative and/or debilitating disorders (Canavan disease (CD; MIM# 271900), cystic fibrosis (CF; MIM# 219700), familial dysautonomia (FD; MIM# 223900), and TSD) [ACOG, 2004]. More recently, the American College of Medical Genetics (ACMG) recommended AJ carrier screening for an additional five neurodegenerative and/or debilitating disorders, including Bloom syndrome (BS; MIM# 210900), Fanconi anemia group C (FA; MIM# 227645), Gaucher disease (GD; MIM# 230800), mucolipidosis type IV (MLIV; MIM# 252650), and Niemann-Pick disease type A (NPD; MIM# 257200) [Gross et al., 2008].
In addition to screening for these nine diseases, many laboratories also test for maple syrup urine disease (MSUD; MIM# 248600) and glycogen storage disease Ia (GSDIa; MIM# 232200), bringing the total number of diseases in these prenatal AJ screening panels to at least 11 with reported carrier frequencies ranging from 0.067 (1 in 15) for GD [Horowitz et al., 1998] to 0.008 (1 in 127) for MLIV [Edelmann et al., 2002]. AJ founder mutations also have been described for lipoamide dehydrogenase deficiency (E3; MIM# 248600) [Hong et al., 2003; Shaag et al., 1999], familial hyperinsulinism (HI; MIM# 256450) [Nestorowicz et al., 1996], nemaline myopathy (NM; MIM# 256030) [Anderson et al., 2004], and Usher syndrome types I (USH1F; MIM# 602083) [Ben-Yosef et al., 2003; Brownstein et al., 2004] and III (USH3; MIM# 276902) [Ness et al., 2003], prompting their recent inclusion into some AJ prenatal screening panels.
Here, we report our experience with carrier screening and prenatal diagnoses for 16 AJ panel disorders, including the rationale and justification for the panel expansion, as well as its acceptance by the screenees. The identified carrier frequencies of the underlying founder mutations in these diseases are based on carrier screening in the New York metropolitan area AJ population, which included a large number of screenees who reported that both parents were 100% AJ. The findings reported here for AJ screening in a large population provides a benchmark for future carrier detection in this and other AJ and ethnic subpopulations.
METHODS
Study Population
Peripheral blood samples were obtained with informed consent from individuals requesting carrier screening from the greater New York metropolitan area from 1996 to 2009. The total screened population was composed of AJ individuals, individuals with unknown or mixed ancestry, and non-AJ individuals most of which had AJ spouses. Among these screenees, a large cohort was identified who reported that both parents were 100% AJ and this population was used to determine AJ carrier frequencies and to calculate residual risk values.
Carrier Screening Assays
Until 2005, all diseases included in our prenatal AJ panel were tested using clinically approved standard polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) and allele-specific oligonucleotide hybridization (ASOH) assays [Dong et al., 2002; Edelmann et al., 2002; Edelmann et al., 2001; Eng and Desnick, 2001; Eng et al., 1997; Kornreich et al., 2004]. In 2005, BS, CD, FA, FD, GD, MLIV, NPD and TSD, and CF were genotyped using the AJP and CF Tag-It™ Mutation Detection Kits (Luminex Molecular Diagnostics, Toronto, ON), respectively, according to the manufacturer’s instructions. Genotypes for these samples were determined using Tag-It™ Data Analysis Software (Luminex Molecular Diagnostics). The number of CFTR mutations included in the CF testing panel increased progressively from six in 1996 (p.W1282X, p.F508del, p.G542X, c.3717+12191C>T (3849+10kb C>T), p.N1303K, and p.I507del) to the current panel of 76 mutations and variants, which includes the 23 ACMG-recommended mutations [Grody et al., 2001; Watson et al., 2004] and the p.D1152H mutation which is prevalent in the AJ population [Kornreich et al., 2004].
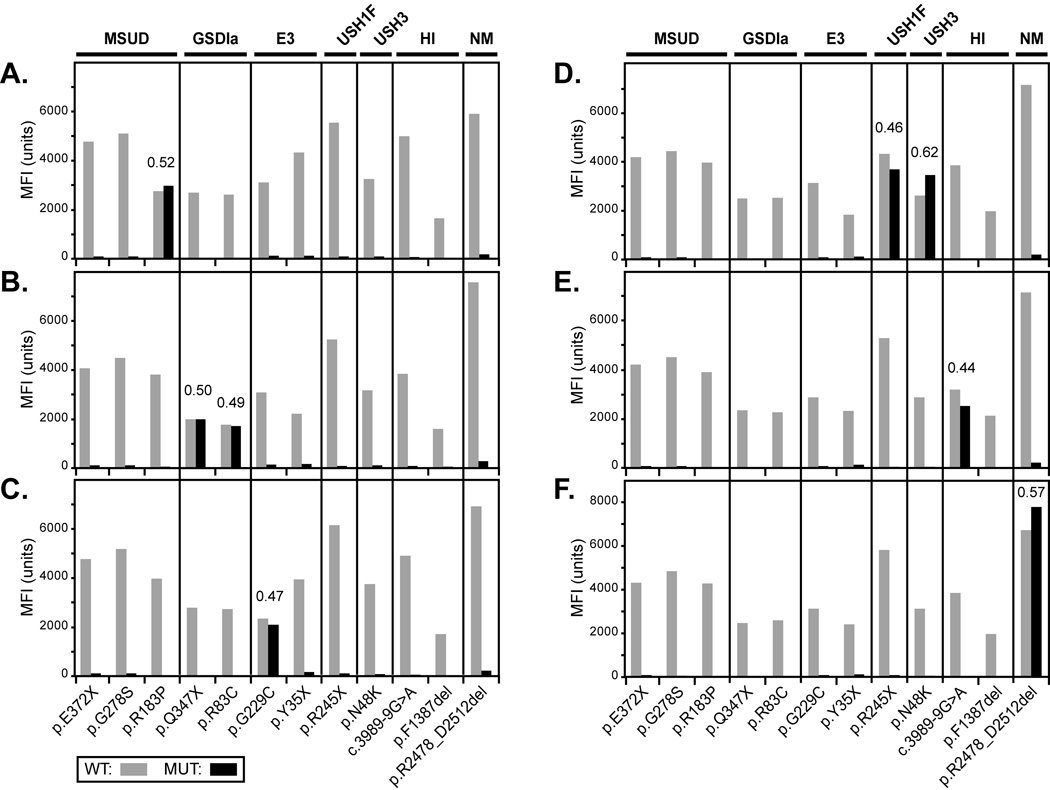
In 2004 and 2005, MSUD and GSDIa, respectively, were added to our AJ panel using clinically approved ASOH assays [Edelmann et al., 2001]. In 2007, they and five additional diseases (E3, HI, NM, USH1F, and USH3) were incorporated into the panel using a multiplex PCR/allele-specific primer extension (ASPE) assay, as described in the Supp. Methods. The 12 specific mutations tested by this assay for these seven diseases are listed in Supp. Table S1, and the PCR and ASPE primer sequences are listed in Supp. Tables S2 and S3, respectively. A graphical output of allelic ratios from representative positive control MSUD, GSDIa, E3, USH1F, USH3, HI and NM samples is shown in Figure 1.
FIGURE 1.
Net MFI data of representative AJ mutation carriers using the MicroPlex™-xTAG™ assay. Results illustrate: (A) a MSUD carrier, (B) a GSDIa compound heterozygote (Coriell ID: NA11468), (C) an E3 carrier, (D) an USH1F and USH3 double-heterozygote, (E) an HI carrier, and (F) a NM carrier. Mutant allelic ratios (see Supporting Methods) are noted above all heterozygous alleles. MFI: mean fluorescence intensity; WT: wild-type; MUT: mutant.
Together, testing for all 16 disorders currently includes 118 total mutations and variants. The GenBank RefSeq and MIM entries for the 16 panel genes are: GD (GBA; NG_009783.1; MIM# 606463), CF (CFTR; NG_016465.1; MIM# 602421), TSD (HEXA; NG_009017.1; MIM# 606869), FD (IKBKAP; NG_008788.1; MIM# 603722), CD (ASPA; NG_008399.1; MIM# 608034), GSDIa (G6PC; NG_011808.1; MIM# 232200), HI (ABCC8; NG_008867.1; MIM# 600509), MLIV (MCOLN1; AF287270.1; MIM# 605248), MSUD (BCKDHB; NG_009775.1; MIM# 248611), FA (FANCC; NG_011707.1; MIM# 227645), E3 (DLD; NG_008045.1; MIM# 238331), NPD (SMPD1; NG_011780.1; MIM# 607608), USH3 (CLRN1; NG_009168.1; MIM# 606397), BS (BLM; NG_007272.1; MIM# 604610), USH1F (PCDH15; NG_009191.1; MIM# 605514), NM (NEB; NG_009382.2; MIM# 161650). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1. Of note, prior to 2005, DNA-based TSD testing was only performed in our laboratory for confirmation of hexosaminidase enzyme analyses. Consequently, for this study, the TSD carrier frequency was determined from individuals screened by the 2005 AJ panel and onward.
RESULTS
Allele and Carrier Frequencies
The carrier frequencies for all screenees and the 100% AJ screenees are summarized in Table 1 and ranged from 0.060 (1 in 16.8; GD) to 0.005 (1 in 186; NM) and from 0.066 (1 in 15.2; GD) to 0.006 (1 in 168; NM), respectively. All disease alleles in both populations were in Hardy-Weinberg equilibrium. On average the 100% AJ carrier frequencies were ~15% higher than those among all screenees, except for CF and NPD where the carrier frequencies were about equal. For the five recently added disorders (E3, HI, NM, USH1F, and USH3), the 100% AJ carrier frequencies ranged from 0.015 (1 in 68; HI) to 0.006 (1 in 168; NM), which resulted in a cumulative carrier frequency of 0.045 (1 in 22).
TABLE 1.
Carrier frequencies for AJ panel mutations
| Total Screened Population |
100% AJ Population |
|||||
|---|---|---|---|---|---|---|
| Disease (Gene): Mutationa | Carriers/ Screenees |
Allele Frequency |
Carrier Frequency | Carriers/ Screenees |
Allele Frequency |
Carrier Frequency |
| GD (GBA): | ||||||
| p.N409S (N370S) | 1,573/30,596 | 0.026 | 0.0514 (1 in 19) | 497/8,425 | 0.029 | 0.0590 (1 in 17) |
| c.84dupG (84GG) | 76/30,596 | 0.0012 | 0.0025 (1 in 403) | 29/8,425 | 0.0017 | 0.0034 (1 in 291) |
| p.R535H (R496H) | 6/2,012 | 0.0015 | 0.0030 (1 in 335) | 2/1,017 | 0.0010 | 0.0020 (1 in 509) |
| p.L483P (L444P) | 42/30,596 | 0.0007 | 0.0014 (1 in 728) | 7/8,425 | 0.0004 | 0.0008 (1 in 1,204) |
| c.115+1G>A (IVS2+1G>A) | 22/30,596 | 0.0004 | 0.0007 (1 in 1,391) | 5/8,425 | 0.0003 | 0.0006 (1 in 1,685) |
| p.V433L (V394L) | 1/2,012 | 0.0002 | 0.0005 (1 in 2,012) | 0/1,017 | 0.000 | - |
| c.1263_1317del (del55bp) | 0/2,012 | 0.000 | - | 0/1,017 | 0.000 | - |
| p.D448H (D409H) | 0/2,012 | 0.000 | - | 0/1,017 | 0.000 | - |
| 0.0595 (1 in 16.8) | 0.0658 (1 in 15.2) | |||||
| CF (CFTR):b | ||||||
| p.W1282X | 503/49,603 | 0.005 | 0.0101 (1 in 99) | 175/8,671 | 0.010 | 0.0202 (1 in 50) |
| p.F508del | 892/49,603 | 0.009 | 0.0180 (1 in 56) | 101/8,671 | 0.006 | 0.0116 (1 in 86) |
| p.D1152Hc | 68/21,854 | 0.0016 | 0.0031 (1 in 321) | 24/4,519 | 0.0027 | 0.0053 (1 in 188) |
| p.G542X | 104/49,603 | 0.0010 | 0.0021 (1 in 477) | 21/8,671 | 0.0012 | 0.0024 (1 in 413) |
| c.3717+12191C>T (3849+10kb C>T) | 61/49,603 | 0.0006 | 0.0012 (1 in 813) | 18/8,671 | 0.0010 | 0.0021 (1 in 482) |
| p.N1303K | 57/49,603 | 0.0006 | 0.0011 (1 in 870) | 15/8,671 | 0.0009 | 0.0017 (1 in 578) |
| p.I148T | 52/21,854 | 0.0012 | 0.0024 (1 in 420) | 2/4,519 | 0.0002 | 0.0004 (1 in 2,260) |
| p.R117Hf | 48/21,854 | 0.0011 | 0.0022 (1 in 911) | 2/4,519 | 0.0002 | 0.0004 (1 in 2,260) |
| c.1585-1G>A (1717-1G>A) | 21/49,603 | 0.0002 | 0.0004 (1 in 2,362) | 2/6,737 | 0.0001 | 0.0003 (1 in 3,369) |
| p.R347Hc | 2/21,854 | 0.00005 | 0.0001 (1 in 10,927) | 1/4,519 | 0.0001 | 0.0002 (1 in 4,519) |
| p.D1270Nc | 10/2,812 | 0.0018 | 0.0036 (1 in 281) | 0/420 | 0.000 | - |
| p.G622Dc | 2/2,812 | 0.0004 | 0.0007 (1 in 1,406) | 0/420 | 0.000 | - |
| p.G551D | 15/49,603 | 0.0002 | 0.0003 (1 in 3,307) | 0/6,737 | 0.000 | - |
| c.2988+1G>A (3120+1G>A) | 4/21,854 | 0.0001 | 0.0002 (1 in 5,464) | 0/4,519 | 0.000 | - |
| p.R334W | 4/21,854 | 0.0001 | 0.0002 (1 in 5,464) | 0/4,519 | 0.000 | - |
| p.R553X | 8/49,603 | 0.0001 | 0.0002 (1 in 6,200) | 0/6,737 | 0.000 | - |
| p.R1162X | 3/21,854 | 0.0001 | 0.0001 (1 in 7,285) | 0/4,519 | 0.000 | - |
| c.1766+1G>A (1898+1G>A) | 2/21,854 | 0.00005 | 0.0001 (1 in 10,927) | 0/4,519 | 0.000 | - |
| p.A455E | 2/21,854 | 0.00005 | 0.0001 (1 in 10,927) | 0/4,519 | 0.000 | - |
| p.R560T | 2/21,854 | 0.00005 | 0.0001 (1 in 10,927) | 0/4,519 | 0.000 | - |
| c.489+1G>T (621+1G>T) | 3/49,603 | 0.00003 | 0.00006 (1 in 16,534) | 0/6,737 | 0.000 | - |
| c.2657+5G>A (2789+5G>A) | 1/21,854 | 0.00002 | 0.00005 (1 in 21,854) | 0/4,519 | 0.000 | - |
| c.579+1G>T (711+1G>T) | 1/21,854 | 0.00002 | 0.00005 (1 in 21,854) | 0/4,519 | 0.000 | - |
| c.3528delC (3659delC) | 1/21,854 | 0.00002 | 0.00005 (1 in 21,854) | 0/4,519 | 0.000 | - |
| p.G85E | 1/21,854 | 0.00002 | 0.00005 (1 in 21,854) | 0/4,519 | 0.000 | - |
| p.I507del | 2/49,603 | 0.00002 | 0.00004 (1 in 24,802) | 0/8,671 | 0.000 | - |
| 0.0442 (1 in 22.6) | 0.0443 (1 in 22.6) | |||||
| TSD (HEXA):e | ||||||
| c.1274_1277dupTATC (1278insTATC) |
55/2,198 | 0.013 | 0.0250 (1 in 40) | 32/1,015 | 0.016 | 0.0315 (1 in 32) |
| c.1421+1G>C (IVS12+1G>C) | 10/2,198 | 0.0023 | 0.0045 (1 in 220) | 5/1,015 | 0.0025 | 0.0049 (1 in 203) |
| p.R247W | 4/2,198 | 0.0009 | 0.0018 (1 in 550) | 1/1,015 | 0.0005 | 0.0010 (1 in 1,015) |
| p.G269S | 2/2,198 | 0.0005 | 0.0009 (1 in 1,099) | 0/1,015 | 0.000 | - |
| c.1073+1G>A (IVS9+1G>A) | 2/2,198 | 0.0005 | 0.0009 (1 in 1,099) | 0/1,015 | 0.000 | - |
| p.R249W | 0/2,198 | 0.000 | - | 0/1,015 | 0.000 | - |
| g.2644_10588del (7.6kb del) | 0/2,198 | 0.000 | - | 0/1,015 | 0.000 | - |
| 0.0314 (1 in 31.9) | 0.0365 (1 in 27.4) | |||||
| FD (IKBKAP):f | ||||||
| c.2204+6T>C (IVS20+6T>C) | 397/14,424 | 0.014 | 0.0275 (1 in 36) | 267/8,207 | 0.016 | 0.0325 (1 in 31) |
| p.R696P | 0/14, 424 | 0.000 | - | 0/5,629 | 0.000 | - |
| 0.0275 (1 in 36.3) | 0.0325 (1 in 30.7) | |||||
| CD (ASPA): | ||||||
| p.E285A | 462/32,072 | 0.007 | 0.0144 (1 in 69) | 134/8,793 | 0.008 | 0.0152 (1 in 66) |
| p.Y231X | 71/32,072 | 0.0011 | 0.0022 (1 in 452) | 26/8,793 | 0.0015 | 0.0030 (1 in 338) |
| p.A305E | 2/32,072 | 0.00003 | 0.0001 (1 in 16,036) | 0/8,793 | 0.000 | - |
| c.433-2A>G | 0/1,820 | 0.000 | - | 0/916 | 0.000 | - |
| 0.0167 (1 in 59.9) | 0.0182 (1 in 55.0) | |||||
| GSDIa (G6PC): | ||||||
| p.R83C | 51/4,228 | 0.006 | 0.0121 (1 in 83) | 47/3,012 | 0.008 | 0.0156 (1 in 64) |
| p.Q347X | 0/4,151 | 0.000 | - | 0/3,012 | 0.000 | - |
| 0.0121 (1 in 82.9) | 0.0156 (1 in 64.1) | |||||
| HI (ABCC8): | ||||||
| c.3989-9G>A (3992-9G>A) | 134/12,886 | 0.005 | 0.0104 (1 in 96) | 91/7,372 | 0.006 | 0.0123 (1 in 81) |
| p.F1387del (delF1388) | 22/12,886 | 0.0009 | 0.0017 (1 in 586) | 18/7,372 | 0.0012 | 0.0024 (1 in 410) |
| 0.0121 (1 in 82.6) | 0.0148 (1 in 67.6) | |||||
| MLIV (MCOLN1):g | ||||||
| c.406-2A>G (IVS3-2A>G) | 91/12,953 | 0.0035 | 0.0070 (1 in 142) | 56/6,648 | 0.0042 | 0.0084 (1 in 119) |
| g.511_6943del (6.4kb del) | 34/12,953 | 0.0013 | 0.0026 (1 in 381) | 19/6,648 | 0.0014 | 0.0029 (1 in 350) |
| 0.0097 (1 in 103.6) | 0.0113 (1 in 88.6) | |||||
| MSUD (BCKDHB):h | ||||||
| p.R183P | 44/6,867 | 0.0032 | 0.0064 (1 in 156) | 43/5,142 | 0.0042 | 0.0084 (1 in 120) |
| p.G278S | 6/6,867 | 0.0004 | 0.0009 (1 in 1154) | 10/5,253 | 0.0010 | 0.0019 (1 in 525) |
| p.E372X | 0/6,867 | 0.000 | - | 0/5,253 | 0.000 | - |
| 0.0073 (1 in 137.3) | 0.0103 (1 in 97.4) | |||||
| FA (FANCC): | ||||||
| c.456+4A>T (IVS4+4A>T) | 173/20,312 | 0.0043 | 0.0085 (1 in 117) | 66/6,620 | 0.005 | 0.0100 (1 in 100) |
| c.67delG (322delG) | 0/1,780 | 0.000 | - | 0/914 | 0.000 | - |
| 0.0085 (1 in 117.4) | 0.0100 (1 in 100.3) | |||||
| E3 (DLD): | ||||||
| p.G229C | 114/12,833 | 0.0044 | 0.0089 (1 in 113) | 67/7,384 | 0.0045 | 0.0091 (1 in 110) |
| p.Y35X | 2/12,833 | 0.0001 | 0.0002 (1 in 6,417) | 2/7,384 | 0.0001 | 0.0003 (1 in 3,692) |
| 0.0090 (1 in 110.6) | 0.0093 (1 in 107.0) | |||||
| NPD (SMPD1):i | ||||||
| p.R498L (R496L) | 146/29,665 | 0.0025 | 0.0049 (1 in 203) | 52/10,709 | 0.0024 | 0.0049 (1 in 206) |
| c.996delC (fsP330) | 74/29,665 | 0.0012 | 0.0025 (1 in 401) | 29/10,709 | 0.0014 | 0.0027 (1 in 369) |
| p. L304P (L302P) | 33/29,665 | 0.0006 | 0.0011 (1 in 899) | 11/10,709 | 0.0005 | 0.0010 (1 in 974) |
| p.R610del (p.R608del) | 12/29,665 | 0.0002 | 0.0004 (1 in 2,472) | 1/9,709 | 0.0001 | 0.0001 (1 in 9,709) |
| 0.0089 (1 in 111.9) | 0.0087 (1 in 115.0) | |||||
| USH3 (CLRN1):j | ||||||
| p.N48K | 99/12,856 | 0.0039 | 0.0077 (1 in 129.9) | 65/7,792 | 0.0042 | 0.0083 (1 in 119.9) |
| BS (BLM):k | ||||||
| c.2207_2212delATCTGAinsTAGAT TC (2281del6/ins7) |
149/20,269 | 0.0037 | 0.0074 (1 in 136.0) | 61/8,158 | 0.0037 | 0.0075 (1 in 133.7) |
| USH1F (PCDH15): | ||||||
| p.R245X | 56/8,700 | 0.0032 | 0.0064 (1 in 155.4) | 35/5,154 | 0.0034 | 0.0068 (1 in 147.3) |
| NM (NEB): | ||||||
| p.R2478_D2512del | 69/12,853 | 0.0027 | 0.0054 (1 in 186.3) | 44/7,386 | 0.0030 | 0.0060 (1 in 167.9) |
The GenBank RefSeq numbers for the 16 panel genes are: GBA (NG_009783.1), CFTR (NG_016465.1), HEXA (NG_009017.1), IKBKAP (NG_008788.1), ASPA (NG_008399.1), G6PC (NG_011808.1), ABCC8 (NG_008867.1), MCOLN1 (AF287270.1), BCKDHB (NG_009775.1), FANCC (NG_011707.1), DLD (NG_008045.1), SMPD1 (NG_011780.1), CLRN1 (NG_009168.1), BLM (NG_007272.1), PCDH15 (NG_009191.1), NEB (NG_009382.2). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1. Common or ‘Legacy’ names for selected mutations are listed in brackets.
Only detected CF mutations are listed; non-pathogenic p.I148T alleles (italicized) were not included in the combined carrier frequency calculation; combined data with [Kornreich et al., 2004].
CFTR mutations not included in the 23 ACMG-recommended CF panel [Grody et al., 2001; Watson et al., 2004].
p.R117H frequency includes all c.1210-12T[5_9] poly-T tract lengths, including an unknown tract length.
The TSD pseudodeficiency alleles (p.R247W and p.R249W; italicized) were not included in the combined carrier frequency calculation.
Combined FD data with [Dong et al., 2002].
Combined MLIV data with [Edelmann et al., 2002].
Combined MSUD data with [Edelmann et al., 2001].
Combined NPD data with [Caggana et al., 1994].
Combined USH3 data with [Ness et al., 2003].
Combined BS data with [Li et al., 1998].
Regarding CF, while six CFTR mutations (p.W1282X, p.F508del, p.D1152H, p.G542X, c.3717+12191C>T, and p.N1303K) accounted for >98% of the total mutations identified among the 100% AJ screenees, they only accounted for ~86% of the mutations identified among all screenees. In addition, whereas p.F508del was the most common mutation identified among all screenees (0.018; 1 in 56), p.W1282X was the most common mutation identified among the 100% AJ screenees with a slightly higher frequency (0.020; 1 in 50). Although the ACMG recommends testing 23 mutations for routine CF carrier screening [Grody et al., 2001; Watson et al., 2004], additional CFTR mutations were identified in 4.5% and 7.0% of all screenees and the 100% AJ screenees, respectively, primarily due to the p.D1152H mutation prevalent among the AJ (1 in 188) [Kornreich et al., 2004].
Residual Risks
All 16 diseases included in the prenatal carrier panel have high detectability in the AJ population (>0.90) with the exception of USH1F, which is estimated to be ≥0.75. These rates are generally accepted for the majority of diseases [DeMarchi et al., 1996; Eng and Desnick, 2001; Eng et al., 1997; Gross et al., 2008; Monaghan et al., 2008], and the detectabilities of the five disorders recently included in the panel were based on previous reports of affected AJ families [Anderson et al., 2004; Ben-Yosef et al., 2003; Brownstein et al., 2004; Hong et al., 2003; Ness et al., 2003; Nestorowicz et al., 1996; Shaag et al., 1999]. Their respective detectability rates were used to determine the residual risk for each disease among the 100% AJ screenees, which are summarized in Table 2. Due to a carrier frequency of 0.066 (1 in 15), GD had the highest residual risk (1 in 281), whereas BS had the lowest (1 in 13,301) when compared to the other disorders. Of the five disorders recently included in the panel, USH1F had the highest residual risk (1 in 585) as a result of its lower detectability (≥0.75) and NM had the lowest (1 in 3,341).
TABLE 2.
Residual risk values for AJ diseases
| Disease | 100% AJ Carrier Frequency |
Detectability | Residual Risk |
Probability of Affected Fetus if Parents pos/nega |
Probability of Affected Fetus if Parents neg/negb |
|---|---|---|---|---|---|
| GD | 1 in 15 | 0.95 | 1 in 281 | 1 in 1,124 | 1 in 3.2 × 105 |
| CF | 1 in 23 | 0.94 | 1 in 368 | 1 in 1,472 | 1 in 5.4 × 105 |
| TSD | 1 in 27 | 0.98 | 1 in 1,301 | 1 in 5,204 | 1 in 6.8 × 106 |
| FD | 1 in 31 | >0.99 | 1 in 3,001 | 1 in 12,004 | 1 in 3.6 × 107 |
| CD | 1 in 55 | >0.97 | 1 in 1,801 | 1 in 7,204 | 1 in 1.3 × 107 |
| GSDIa | 1 in 64 | 0.95 | 1 in 1,261 | 1 in 5,044 | 1 in 6.4 × 106 |
| HI | 1 in 68 | 0.90 | 1 in 671 | 1 in 2,684 | 1 in 1.8 × 106 |
| MLIV | 1 in 89 | 0.95 | 1 in 1,761 | 1 in 7,044 | 1 in 1.2 × 107 |
| MSUD | 1 in 97 | 0.95 | 1 in 1,921 | 1 in 7,684 | 1 in 1.5 × 107 |
| FA | 1 in 100 | 0.99 | 1 in 9,901 | 1 in 39,604 | 1 in 3.9 × 108 |
| E3 | 1 in 107 | >0.95 | 1 in 2,121 | 1 in 8,484 | 1 in 1.8 × 107 |
| NPD | 1 in 115 | 0.97 | 1 in 3,801 | 1 in 15,204 | 1 in 5.8 × 107 |
| USH3 | 1 in 120 | >0.95 | 1 in 2,381 | 1 in 9,524 | 1 in 2.3 × 107 |
| BS | 1 in 134 | 0.99 | 1 in 13,301 | 1 in 53,204 | 1 in 7.1 × 108 |
| USH1F | 1 in 147 | ≥0.75 | 1 in 585 | 1 in 2,340 | 1 in 1.4 × 106 |
| NM | 1 in 168 | >0.95 | 1 in 3,341 | 1 in 13,364 | 1 in 4.5 × 107 |
One parent is positive and one parent is negative by carrier screening.
Both parents are negative by carrier screening.
Table 2 also lists the residual risk values of having an affected fetus when one parent tests positive and the other negative (i.e., positive/negative) and when both parents test negative (i.e., negative/negative) for each of the 16 AJ disorders. In the presence of a positive/negative or negative/negative parental test result, GD had the highest residual risk for an affected fetus (1 in 1,124 or 1 in 3.2 × 105, respectively), whereas BS had the lowest risk (1 in 53,204 or 1 in 7.1 × 108, respectively).
Combined Carrier Frequencies of AJ Testing Panels
The combined carrier frequencies for the AJ carrier screening disorders are summarized in Table 3. As expected, the combined carrier frequencies for all screenees and the 100% AJ screenees increased with the number of diseases tested. The combined carrier frequencies for the 16 disorders among all screenees and the 100% AJ screenees were 0.274 (1 in 3.7) and 0.306 (1 in 3.3), respectively.
TABLE 3.
Chronological combined carrier frequencies of AJ panel disorders
| Combined Carrier Frequency |
||||
|---|---|---|---|---|
| Yeara | Disorders Addedb | Number of Disorders in Panel |
Total Screenees | 100% AJ Screenees |
| 1975 | TSD | 1 | 0.0314 (1 in 31.9) | 0.0365 (1 in 27.4) |
| 1992 | GD, CF | 3 | 0.1351 (1 in 7.4) | 0.1648 (1 in 6.8) |
| 1996 | CD, NPD | 5 | 0.1607 (1 in 6.2) | 0.1735 (1 in 5.8) |
| 1999 | BS, FA | 7 | 0.1766 (1 in 5.7) | 0.1909 (1 in 5.2) |
| 2001 | FD | 8 | 0.2041 (1 in 4.9) | 0.2235 (1 in 4.5) |
| 2002 | MLIV | 9 | 0.2137 (1 in 4.7) | 0.2348 (1 in 4.3) |
| 2004 | MSUD | 10 | 0.2210 (1 in 4.5) | 0.2450 (1 in 4.1) |
| 2005 | GSDIa | 11 | 0.2331 (1 in 4.3) | 0.2606 (1 in 3.8) |
| 2007 | E3, HI, USH1F, USH3, NM | 16 | 0.2737 (1 in 3.7) | 0.3058 (1 in 3.3) |
Year that disorders were incorporated into panel.
Routine TSD carrier screening was performed by enzyme analysis until 2005.
Double- and Triple-Heterozygote Carrier Frequencies
The AJ double-heterozygote frequencies are summarized in Supp. Table S4. Of note, 1 in 21 AJ individuals was found to be a carrier for two of the 16 diseases, consistent with the predicted frequency of ~1 in 24 when using the full testing panel. Together, GD and CF mutations were the most commonly identified alleles among double-heterozygous AJ individuals with an observed frequency (1 in 400) consistent with their predicted frequency of ~1 in 343 (Supp. Table S5). In addition, since 2002, seven triple-heterozygous AJ individuals have been identified by routine screening, resulting in an observed frequency of 0.0018 (~1 in 561). Each of these individuals harbored unique disease mutation combinations (GD/CF/NPD, GD/CF/BS, GD/TSD/FD, GD/FD/CD, CF/FD/FA, CF/GSDIa/MSUD, and TSD/CD/FA).
Panel Selection among AJ Screenees
A random subset of AJ screenees was monitored to evaluate the disorders selected for carrier testing and these results are summarized in Table 4. Among 466 AJ individuals who underwent pre-screening genetic counseling between 2008 and 2009, the majority (82%) selected a testing panel that included all 16 disorders. Of those who did not choose all 16 disorders, the majority chose the full panel without CF because they had previously undergone CF testing and/or had testing performed elsewhere. Not including CF, ~96% of AJ individuals chose to include all available disorders and these results were consistent whether they underwent genetic counseling at our center or elsewhere. Importantly, less than 2% of the AJ population chose not to include either USH1F or the five recently added disorders (E3, HI, NM, USH1F, and USH3) in their panel, suggesting that the AJ population is strongly in favor of including all 16 disorders in prenatal testing panels.
TABLE 4.
Panel selection among AJ screenees
| Total Population |
MSSMa |
Non-MSSMb |
||||
|---|---|---|---|---|---|---|
| Panel | n | Frequency | n | Frequency | n | Frequency |
| Full Panel (16 disorders) | 383 | 0.822 | 248 | 0.855 | 135 | 0.767 |
| Panel without CF | 68 | 0.146 | 37 | 0.128 | 31 | 0.176 |
| Panel without USH1F | 8 | 0.017 | 2 | 0.007 | 6 | 0.034 |
| Panel without E3, USH1F, USH3, HI and NM | 7 | 0.015 | 3 | 0.010 | 4 | 0.023 |
| Total | 466 | 1.0 | 290 | 1.0 | 176 | 1.0 |
n: number of AJ individuals.
AJ cohort who underwent genetic counseling at Mount Sinai School of Medicine.
AJ cohort who did not undergo genetic counseling at Mount Sinai School of Medicine.
Prenatal Testing
Prenatal diagnostic testing has been performed at our center for all 16 AJ disorders and the cumulative results since 1996 are summarized in Table 5. Among all DNA-based prenatal diagnoses (n = 574), the fetuses were diagnosed as non-carriers (193; 34%), carriers (274; 48%), or affected (107; 19%). These outcomes significantly deviated from the expected Mendelian ratios (χ2 = 26.9, 2 df) and were due to the fact that prenatal diagnoses were performed for pregnancies in which one partner was a known mutation carrier and the other: 1) was unavailable for testing and had an unknown mutation status, 2) was a carrier by enzyme for TSD but was negative for the commonly screened mutations, or 3) was negative for the commonly screened GD causing mutations but was being tested prenatally by enzyme analysis.
TABLE 5.
Prenatal testing results of AJ panel disorders
| All Monitored Pregnancies |
Pregnancies with Both Parents DNA-Confirmeda |
Pregnancies with One Parent DNA-Confirmedb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disorder | Total | Non- carriers |
Carriers | Affected | Total | Non- carriers |
Carriers | Affected | Total | Non- carriers |
Carriers | Affected |
| TSD | 157 | 50 | 76 | 31c | 102 | 20 | 51 | 31c | 55 | 30 | 25 | 0 |
| CF | 151 | 50 | 68 | 33d | 134 | 33 | 66 | 33d | 17 | 15 | 2 | 0 |
| GD | 144 | 58 | 67 | 19e,f | 65 | 17 | 29 | 19e,f | 79 | 41 | 38 | 0 |
| FD | 40 | 8 | 22 | 10 | 39 | 8 | 21 | 10 | 1 | 0 | 1 | 0 |
| CD | 36 | 11 | 19 | 6 | 28 | 7 | 15 | 6 | 8 | 4 | 4 | 0 |
| NPD | 17 | 3 | 10 | 4 | 17 | 3 | 10 | 4 | 0 | 0 | 0 | 0 |
| MLIV | 8 | 3 | 5 | 0 | 8 | 3 | 5 | 0 | 0 | 0 | 0 | 0 |
| GSDIa | 4 | 1 | 1 | 2 | 4 | 1 | 1 | 2 | 0 | 0 | 0 | 0 |
| E3 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 1 | 0 |
| MSUD | 3 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| HI | 2 | 1 | 0 | 1f | 2 | 1 | 0 | 1f | 0 | 0 | 0 | 0 |
| USH3 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 |
| USH1F | 2 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| NM | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 |
| FA | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| BS | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Total | 574 | 193 | 274 | 107 | 404 | 97 | 200 | 107 | 170 | 96 | 74 | 0 |
| Frequency | 1.0 | 0.336 | 0.477 | 0.186 | 1.0 | 0.240 | 0.495 | 0.265 | 1.0 | 0.565 | 0.435 | 0.000 |
Both parents confirmed carriers by DNA analysis.
One parent a confirmed carrier by DNA analysis.
Includes one c.[1274_1277dupTATC] + p.[R247W] compound heterozygote (see Results and Discussion).
Includes p.[W1282X] + c.[1210-12T[5]], p.[F508del] + c.[1210-12T[5]], p.[G542X] + [R117H] (without c.1210-12T[5]), and p.[N1303K] + [R117H] (without c.1210-12T[5]) compound heterozygotes (see Results and Discussion).
Includes fourteen p.N370S homozygotes, one p.R496H homozygote, and three p.[N370S] + c.[84dupG] and one p.[N370S] + [R496H] compound heterozygotes (see Results and Discussion).
Includes one fetus with p.[N370S] + [N370S] and c.[3989-9G>A] + p.[F1387del] affected with both GD and HI (see Results and Discussion).
Among the 404 DNA confirmed at-risk pregnancies, 97 (24%), 200 (50%), and 107 (27%) were determined by DNA analysis to be non-carriers, carriers, and affected, respectively, the distribution being consistent with the expected Mendelian ratios (χ2 = 0.53, 2 df). Of the 31 TSD fetuses harboring two mutations, one was compound heterozygous for the HEXA c.1274_1277dupTATC null mutation and the p.R247W pseudodeficiency allele; however, after counseling the parents elected to continue the pregnancy. Genotypes with highly variable phenotypes also were observed in four CF pregnancies, including p.[W1282X] + c.[1210-12T[5]], p.[F508del] + c.[1210-12T[5]], p.[G542X] + [R117H] (without c.1210-12T[5]), and p.[N1303K] + [R117H] (without c.1210-12T[5]) compound heterozygotes. Among these, two sets of parents continued the pregnancies (p.[F508del] + c.[1210-12T[5]] and p.[N1303K] + [R117H]), whereas the parents of the p.[W1282X] + c.[1210-12T[5]] and p.[G542X] + [R117H] fetuses elected termination. Additionally, among the 19 fetuses predicted to be affected with GD, 14 were found to be p.[N370S] + [N370S], a mild phenotype [Balwani et al., 2010], and all but one couple elected to continue these pregnancies.
Among the 170 pregnancies with only a single DNA confirmed parent, 96 (56%) and 74 (44%) were determined by DNA analysis to be non-carriers and carriers, respectively. The majority (n = 134; 79%) of these monitored pregnancies were for GD (n = 79) and TSD (n = 55), in part due to their higher carrier frequencies (1 in 15 and 1 in 27, respectively). These pregnancies were referred to our center which offers prenatal enzyme and/or DNA diagnoses for these diseases. No affected pregnancies were detected in these cohorts. The outcomes from the combined cohort deviated significantly from the expected Mendelian ratios (χ2 = 111.3, 2 df), suggesting that most of the unconfirmed parents of these pregnancies were not mutation carriers.
Interestingly, one fetus was identified as affected with both GD and HI as both parents were double-heterozygotes (GBA: p.[N370S] + ABCC8: c.[3989-9G>A], and GBA: p.[N370S] + ABCC8: p.[F1387del]). Based on the 100% AJ carrier frequencies, the predicted frequency of GD/HI double-heterozygous individuals was 1 in 1,027, and the frequency of an at-risk carrier couple for both diseases was lower than 1 in 106.
DISCUSSION
AJ Carrier Screening and Panel Expansion
Almost 40 years ago, prenatal carrier screening for TSD in the AJ community became the prototype for the prevention of severe recessive genetic diseases. The paradigm of educating the community, offering testing options, and genetic counseling was readily accepted in the AJ community and has expanded to other ethnic, demographic, and racial groups for the option to prevent seriously debilitating diseases of childhood in at-risk populations (e.g., Dor Yeshorim in the ultra orthodox community [Ekstein and Katzenstein, 2001], Druze [Falik-Zaccai et al., 2008] and Saudi Arabian [Al Sulaiman et al., 2008] screening programs, thalassemia [Cao et al., 2002] and the hemoglobinopathies in the Mediterranean [Patrinos et al., 2005]). As the genes and founder/common mutations for other severely debilitating and neurodegenerative diseases prevalent in the AJ population were identified, additional diseases were included in the AJ prenatal screening panel, largely driven by demand from the AJ community itself, particularly from parents who were screened for the more frequent disorders but who had an affected child with a less frequent disease not included in the panel. Suffice it to say, AJ parents want to test for all possible debilitating/neurodegenerative disorders that can affect their children. The success of this prevention strategy in the AJ community has been remarkable. Today, the birth of affected children with these diseases is rare [Lerner, 2009] as the screening programs have essentially become ‘standard of care’ by guidelines and recommendations of the ACOG [ACOG, 2004; 2009] and ACMG [Gross et al., 2008; Monaghan et al., 2008].
We recently increased our prenatal AJ carrier screening panel from 11 to 16 disorders prevalent in the AJ population, as parents of affected patients sought testing panel expansion to prevent the birth of children with these diseases. Although there is debate regarding the expansion of prenatal AJ panels [Fares et al., 2008; Gross et al., 2008], there is a compelling rationale for including E3, USH1F, USH3, HI and NM when screening the AJ population. For example, E3 can be a severe neonatal disorder with a fatal outcome. Although it is considered rare, it has been reported in the North American AJ population and it may be under-diagnosed given its clinical presentation which is similar to ketotic hypoglycemia [Sansaricq et al., 2006]. Awareness of the possibility of fatal decompensation could lead to life-saving treatment. However, discussion of the phenotypic variability of p.G229C homozygotes should be included during prenatal genetic counseling. Regarding USH, the severity of profound prelingual hearing loss, vestibular areflexia, prepubertal onset of retinitis pigmentosa, and the occurrence of an AJ founder mutation in USH1F make it a good screening candidate. Although USH3 has a much more variable and later onset than USH1, screening for the CLRN1 p.N48K mutation in the AJ population would inform carrier families of the progressive nature of USH3 and assist in distinguishing it from other atypical forms of USH and non-syndromic deafness. HI is a good candidate for AJ carrier screening given its severe neonatal phenotype and a carrier frequency (0.015; 1 in 68) similar to that observed in many other disorders already included in AJ panels. NM is a variable and genetically heterogeneous disorder; however, the typical form associated with NEB mutations is characterized by infantile onset and profound weakness of facial, bulbar, and respiratory muscles and neck flexors. Although the disease is rare, screening for the p.R2478_D2512del mutation could reduce the occurrence of this devastating disorder in the AJ population.
Further support for inclusion of E3, USH1F, USH3, HI and NM in testing panels comes from the AJ community itself. By monitoring panel selection among AJ screenees, our study determined that following standard pre-screening genetic counseling, the vast majority (~95%) of AJ individuals opted for the expanded testing panel, even after counseling about the lower detectability and residual risk of USH1F. Of note, in addition to information on USH1F, prenatal carrier counseling at our center typically includes a discussion of recessive disease inheritance, carrier frequencies of each disorder, an overview of the clinical features of each disease including those with variable phenotypes, detectability, as well as information on prenatal testing and preimplantation genetic diagnosis availability. The rare possibility of detecting homozygosity – particularly for GD – during carrier screening is also discussed. Taken together, these data indicate that the local AJ community is supportive of carrier screening expansion, including disorders with lower carrier frequencies and/or detectability.
DNA-Based Testing in the AJ Population
Coinciding with the expansion of AJ carrier screening panels has been the continued development and clinical evaluation of multiplexed genotyping platforms [Edelmann et al., 2004; Fares et al., 2008; Kalman et al., 2009; Schrijver et al., 2007; Strom et al., 2004; Strom et al., 2005]. Although the current panel of 16 disorders utilizes commercial assays [Strom et al., 2005; Strom et al., 2006], to facilitate carrier testing of the recently added diseases (E3, USH1F, USH3, HI and NM), we designed a novel multiplexed bead-based assay which simultaneously genotyped their seven mutations and an additional five AJ mutations which cause MSUD and GSDIa.
In addition to targeted mutation analyses, full gene sequencing is increasingly being used to identify rare mutations in some AJ panel disorders (e.g., TSD, CF), particularly for non-AJ or mixed-ethnicity couples. Although this testing strategy offers an increased mutation detection sensitivity, this is offset by the additional cost, labor and turnaround time, and the concerning possibility of identifying sequence variants of unknown significance (e.g., synonymous and non-coding variants). However, many of the AJ individuals surveyed at our institution said they would be inclined to pursue full gene sequencing if they were found to be a carrier of a particular disorder and their partner was not, particularly if their partner was not of AJ descent (unpublished observations).
Regarding cost and turnaround time of full gene sequencing, it is possible that customized resequencing microarrays [Lebet et al., 2008; Waldmuller et al., 2008] and next-generation sequencing platforms [ten Bosch and Grody, 2008; Voelkerding et al., 2009] will decrease in price in the near future, which could enable more thorough and practical clinical sequencing-based assays. Of note, the size of the genes responsible for the 16 disorders in the AJ panel range from four exons (USH3) to 150 exons (NM) (average: 26 exons; median: 14 exons), underscoring the challenge and time required to perform traditional capillary-based sequencing for these diseases. Additionally, forthcoming clinical sequencing platforms should also be able to detect small and large deletions/duplications in conjunction with single nucleotide mutations. This is particularly relevant given that seven of the 16 genes included in the current AJ panel (GBA, G6PC, ABCC8, MCOLN1, DLD, PCDH15, NEB) lie in genomic regions with variable copy-number based on the Database of Genomic Variants (http://projects.tcag.ca/variation/) [Iafrate et al., 2004].
AJ Panel Carrier Frequencies and Residual Risks
The identified carrier frequencies for the 16 disorders among the 100% AJ screenees from the New York metropolitan area were compared to previously reported frequencies from other AJ cohorts (Table 6). This is the first comprehensive study of all 16 disorders involving large numbers of individuals who reported 100% AJ ethnicity, and to our knowledge, the first reported HI carrier frequency of the ABCC8/SUR1 c.3989-9G>A and p.F1387del mutations in the AJ population (0.015; 1 in 68). Excluding TSD, ~3,000 to 11,000 100% AJ individuals were tested for each of the 16 AJ panel disorders to ascertain their allele frequencies, carrier frequencies, and residual risk values.
TABLE 6.
Carrier frequencies of the 16 disorders included in our prenatal AJ panel
| Disease | New York 100% AJ Carrier Frequency (n) |
Published AJ Carrier Frequencya (n) |
Reference |
|---|---|---|---|
| Gaucher Disease | 1 in 15 (8,425) |
1 in 12 (1,364) 1 in 17 (1,208) 1 in 17 (3,336) 1 in 18 (3,764) |
[DeMarchi et al., 1996] [Horowitz et al., 1998] [Fares et al., 2008] [Eng et al., 1997] |
| Cystic Fibrosis | 1 in 23 (8,671) |
1 in 23 (>110,000) 1 in 25 (3,792) 1 in 26 (6,076) |
[Kornreich et al., 2004] [Eng et al., 1997] [Abeliovich et al., 1996] |
| Tay-Sachs Disease | 1 in 27 (1,015) |
1 in 21 (2,824) 1 in 26 (11,008) |
[Eng et al., 1997] [Broide et al., 1993] |
| Familial Dysautonomia | 1 in 31 (8,207) |
1 in 29 (3,246) 1 in 32 (2,518) 1 in 32 (1,100) |
[Fares et al., 2008] [Dong et al., 2002] [Lehavi et al., 2003] |
| Canavan Disease | 1 in 55 (8,793) |
1 in 37 (5,414) 1 in 57 (1,423) 1 in 59 (879) |
[Fares et al., 2008] [Feigenbaum et al., 2004] [Elpeleg et al., 1994] |
| Glycogen Storage Disease Ia | 1 in 64 (3,012) |
1 in 71 (20,719) | [Ekstein et al., 2004] |
| Familial Hyperinsulinism | 1 in 68 (7,372) |
- | - |
| Mucolipidosis Type IV | 1 in 89 (6,648) |
1 in 68 (2,161) 1 in 97 (66,749) 1 in 111 (2,000) 1 in 127 (2,029) |
[Fares et al., 2008] [Bach et al., 2005] [Bargal et al., 2001] [Edelmann et al., 2002] |
| Maple Syrup Urine Disease | 1 in 97 (5,253) |
1 in 113 (1,014) | [Edelmann et al., 2001] |
| Fanconi Anemia Group C | 1 in 100 (6,620) |
1 in 77 (2,782) 1 in 89 (3,104) 1 in 92 (4,029) |
[Fares et al., 2008] [Verlander et al., 1995] [Peleg et al., 2002] |
| Lipoamide Dehydrogenase Deficiency | 1 in 107 (7,384) |
1 in 94 (845) | [Shaag et al., 1999] |
| Niemann-Pick Disease | 1 in 115 (10,709) |
1 in 90 (1,000) 1 in 103 (1,960) |
[Caggana et al., 1994] [Fares et al., 2008] |
| Usher Syndrome Type III | 1 in 120 (7,792) |
1 in 140 (419) | [Ness et al., 2003] |
| Bloom Syndrome | 1 in 134 (8,158) |
1 in 101 (1,613) 1 in 107 (1,491) 1 in 111 (4,001) 1 in 157 (2,680) |
[Shahrabani-Gargir et al., 1998] [Li et al., 1998] [Peleg et al., 2002] [Fares et al., 2008] |
| Usher Syndrome Type IFb | 1 in 147 (5,154) |
1 in 101 (505) 1 in 126 (379) |
[Brownstein et al., 2004] [Ben-Yosef et al., 2003] |
| Nemaline Myopathy | 1 in 168 (7,386) |
1 in 108 (4,090) | [Anderson et al., 2004] |
Previously reported carrier frequencies include American and/or Israeli AJ cohorts; some studies did not test all known disease mutations.
Inclusion of USH1F in the panel is optional due to its detectability of ≥0.75 (see Results and Discussion).
Although several of the disorders had similar frequencies to those previously reported from other AJ cohorts (GD, CF, TSD, FD, CD, GSDIa, MSUD, FA, E3), some diseases warrant further comment. For example, our USH3 carrier frequency (0.0083; 1 in 120) was higher than previously reported (0.0071; 1 in 140) [Ness et al., 2003]; however, this was a limited study of 419 individuals. Assuming Hardy-Weinberg equilibrium, our updated p.N48K carrier frequency from a much larger cohort predicts an USH3 prevalence of ~2 per 100,000 AJ individuals. If USH3 accounts for ~40% of all USH subtypes, the predicted overall prevalence of USH in the AJ population is 4.4 per 100,000, which is within the prevalence estimates of USH in the general population [Petit, 2001].
The NPD carrier frequency observed among our AJ cohort (0.0087; 1 in 115) was lower than previously reported [Caggana et al., 1994; Schuchman and Miranda, 1997]. This was presumably due to differences in sample sizes; however, our larger AJ cohort (n = 10,709) is likely to be more indicative of the actual frequency. In addition, this NPD carrier frequency is similar to that (0.008; 1 in 125) reported in a large series of AJ and non-AJ screenees [Strom et al., 2004]. We also detected a BS carrier frequency (0.0075; 1 in 134) slightly lower than previously reported [Li et al., 1998; Peleg et al., 2002; Shahrabani-Gargir et al., 1998], possibly as a result of random differences between sample populations. The NM carrier frequency identified in our AJ cohort (0.0060; 1 in 168) was lower than previously reported among AJ enrolled in the Dor Yeshorim screening program (0.009; 1 in 108) [Anderson et al., 2004]. Although the reason for this discrepancy is not clear, it may be due to the Dor Yeshorim cohort being primarily composed of Hasidic AJ from New York and Israel. Frequency differences between these AJ subpopulations previously have been reported for MCOLN1 mutations [Edelmann et al., 2002].
Usher Syndrome Type I and Detectability
The USH1F PCDH15 p.R245X mutation previously was identified in the majority of USH1F AJ families while a second putative PCDH15 mutation, p.M1853L, was identified in compound heterozygosity with p.R245X in a single USH1F patient [Ben-Yosef et al., 2003]. We tested 802 AJ individuals and identified five p.M1853L carriers, resulting in a carrier frequency of 0.006 (1 in 160). Combined with p.R245X, this yielded an AJ carrier frequency of 0.0128 (1 in 77) and predicted an USH1F prevalence much higher than the observed general population prevalence [Petit, 2001]. Together, the high frequency of the p.M1853L variant in our AJ cohort, the lack of homozygous p.M1853L among affected USH1 individuals, and the fact that this allele was identified in non-AJ control subjects [Ben-Yosef et al., 2003], indicated that p.M1853L represents a rare non-synonymous polymorphism and that including it in our prenatal AJ panel was not warranted.
Importantly, although the p.R245X mutation was estimated to account for only ≥75% of the reported pathogenic USH1F alleles [Ben-Yosef et al., 2003], the detectability is likely higher given that some of the affected AJ individuals who did not harbor the screened PCDH15 mutations may have had a different USH subtype. For example, two of four families diagnosed with USH without the screened PCDH15 mutations were actually found to carry USH1B-causing MYO7A (MIM# 276903) mutations [Ben-Yosef et al., 2003]. Nevertheless, using the detectability of ≥75% confers a residual risk of 1 in 585 in non-carrier AJ individuals. Thus, genetic counseling to discuss the reduced detectability and residual risk of USH1F carriers prior to testing is strongly recommended.
Identification of Homozygotes by Carrier Screening
The penetrance of the mutations in the 16 disorder AJ panel are generally very high; however, individuals apparently homozygous for specific sequence variants have been identified on rare occasions by routine carrier screening. For example, the high frequency of GD mutations in the AJ population and the variable phenotype of the p.N370S mutation resulted in the identification of both p.N370S homozygotes and compound heterozygotes during carrier screening [Balwani et al., 2010]. However, almost all asymptomatic GD homozygotes serendipitously diagnosed by prenatal carrier screening actually had disease manifestations following careful clinical assessment [Balwani et al., 2010]. In addition, routine carrier screening also identified an adult homozygous for the E3 causing DLD p.G229C mutation, which was confirmed by sequencing with independent PCR primers. Although homozygosity for this mutation typically is associated with a milder form of the disease, significant childhood morbidity and mortality is observed among p.G229C homozygotes [Hong et al., 2003; Sansaricq et al., 2006; Shaag et al., 1999]. Subsequent evaluation revealed four lifetime episodes of exertion fatigue and an unverified previous diagnosis of hypoglycemia; however, clinical assessment of the patient was essentially normal, including all neurological and biochemical laboratory analyses. Although the reason for the favorable clinical course in this patient is unclear, it is possible that other subunits of the associated dehydrogenase complexes modified the p.G229C-mediated enzyme deficiency.
Additionally, one USH3 p.N48K homozygous adult was identified that, after further clinical evaluation, was found to have retinitis pigmentosa. Carrier screening also identified three individuals whose CF tests failed to generate primer extension signals for the pathogenic p.R75X (c.223C>T) allele. Subsequent sequence analysis of exon 3 revealed homozygosity for a neighboring variant not included in our CF mutation panel, p.R75Q (c.224G>A; rs1800076), which is an allele of uncertain clinical significance [Cohn et al., 2005]. Further evaluation of one of the homozygous adults revealed an unremarkable medical history, suggesting that this CFTR variant likely represents a benign, non-synonymous polymorphism. Together, these data indicate that routine carrier screening for recessive disorders can identify individuals homozygous for pathogenic mutations with previously unrecognized clinical manifestations, as well as rare sequence variants of unknown clinical significance that interfere with molecular testing assays.
Prenatal Testing
Recently, there has been discussion regarding the inclusion of disorders with variable expressivity in AJ testing panels, specifically type I GD [Zuckerman et al., 2007]. The most common GBA mutation among the AJ is p.N370S, which precludes neurological disease and may result in a range of severity in affected patients including onset in childhood to a mild to moderate disease course depending on the second mutation. Although the recent ACMG Practice Guidelines recommend GD carrier screening for the AJ population [Gross et al., 2008], some have argued against GD screening based on the availability of effective treatment, the difficultly in predicting disease severity, and the assumption that about two-thirds of p.N370S homozygotes are asymptomatic and never come to medical attention. However, as noted above, based on prenatal carrier screening, most young adult homozygotes that were detected serendipitously and evaluated at our center were found to have disease manifestations [Balwani et al., 2010]. Although GD is one of the more commonly tested disorders in our prenatal panel (~25% of all prenatal tests), it is notable that the vast majority (93%) of carrier couples with p.N370S homozygous pregnancies elected to continue their pregnancies. Similarly, couples with pregnancies that predicted non-pathogenic or variable TSD and CF phenotypes have been identified and counseled at our institution and most couples continued these pregnancies to term.
Conclusions
These studies describe the first comprehensive carrier frequencies and residual risks from a large AJ cohort tested for 16 diseases prevalent in the AJ population. These data can serve as a reference for future carrier screening and genetic counseling in the AJ population. Of note, the recently added disorders (E3, USH1F, USH3, HI and NM) had a cumulative carrier frequency of 1 in 22, supporting their inclusion in testing panels. Importantly, panel selection data among screenees indicated that the AJ population is very supportive of including these disorders in AJ screening panels, regardless of their carrier frequencies and/or detectability. The acceptance of prenatal screening offers insights into the future potential for whole exome or genome sequencing, or a ‘mutation chip’ of all known mutations causing seriously debilitating diseases of childhood. With the current panel of 16 disorders, approximately 1 in 3.3 AJ individuals will be a carrier for one of these diseases and as high as 1 in 24 for two diseases. Thus, simultaneous screening of both partners, in conjunction with pre-screening genetic counseling, is recommended when testing for this panel of disorders in the AJ population. This may reduce the anxiety when one partner tests positive, and is particularly relevant as diseases are added to the AJ panel and the frequency of being a carrier increases.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by a research grant (1 R01 HG006440) and a grant (5 M01 RR00071) from the Division of Research Resources for the Mount Sinai General Clinical Research Center, both from the National Institutes of Health. The authors thank the Mount Sinai Genetic Testing Laboratory technologists for their performance of many thousands of carrier screening and prenatal tests, and Erin Carney for assistance with data collection.
Mount Sinai School of Medicine has licensed two drug therapies to the Genzyme Corporation; Mount Sinai, Mount Sinai’s Department of Genetics and Genomic Sciences, the Chairman of the Department of Genetics and Genomic Sciences and other faculty members in the department receive royalties from the licensed drug for Fabry disease.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
REFERENCES
- Abeliovich D, Quint A, Weinberg N, Verchezon G, Lerer I, Ekstein J, Rubinstein E. Cystic fibrosis heterozygote screening in the Orthodox Community of Ashkenazi Jews: the Dor Yesharim approach and heterozygote frequency. Eur J Hum Genet. 1996;4:338–341. doi: 10.1159/000472229. [DOI] [PubMed] [Google Scholar]
- Al Sulaiman A, Suliman A, Al Mishari M, Al Sawadi A, Owaidah TM. Knowledge and attitude toward the hemoglobinopathies premarital screening program in Saudi Arabia: population-based survey. Hemoglobin. 2008;32:531–538. doi: 10.1080/03630260802508384. [DOI] [PubMed] [Google Scholar]
- American College of Obstetricians and Gynecologists. ACOG committee opinion. Number 298, August 2004. Prenatal and preconceptional carrier screening for genetic diseases in individuals of Eastern European Jewish descent. Obstet Gynecol. 2004;104:425–428. doi: 10.1097/00006250-200408000-00050. [DOI] [PubMed] [Google Scholar]
- American College of Obstetricians and Gynecologists. ACOG Committee Opinion No. 442: Preconception and prenatal carrier screening for genetic diseases in individuals of Eastern European Jewish descent. Obstet Gynecol. 2009;114:950–953. doi: 10.1097/AOG.0b013e3181bd12f4. [DOI] [PubMed] [Google Scholar]
- Anderson SL, Ekstein J, Donnelly MC, Keefe EM, Toto NR, LeVoci LA, Rubin BY. Nemaline myopathy in the Ashkenazi Jewish population is caused by a deletion in the nebulin gene. Hum Genet. 2004;115:185–190. doi: 10.1007/s00439-004-1140-8. [DOI] [PubMed] [Google Scholar]
- Bach G, Webb MB, Bargal R, Zeigler M, Ekstein J. The frequency of mucolipidosis type IV in the Ashkenazi Jewish population and the identification of 3 novel MCOLN1 mutations. Hum Mutat. 2005;26:591. doi: 10.1002/humu.9385. [DOI] [PubMed] [Google Scholar]
- Balwani M, Fuerstman L, Kornreich R, Edelmann L, Desnick RJ. Type 1 Gaucher Disease: Significant disease manifestations in 'asymptomatic' homozygotes. Arch Intern Med. 2010 doi: 10.1001/archinternmed.2010.302. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargal R, Avidan N, Olender T, Ben Asher E, Zeigler M, Raas-Rothschild A, Frumkin A, Ben-Yoseph O, Friedlender Y, Lancet D and others. Mucolipidosis type IV: novel MCOLN1 mutations in Jewish and non-Jewish patients and the frequency of the disease in the Ashkenazi Jewish population. Hum Mutat. 2001;17:397–402. doi: 10.1002/humu.1115. [DOI] [PubMed] [Google Scholar]
- Ben-Yosef T, Ness SL, Madeo AC, Bar-Lev A, Wolfman JH, Ahmed ZM, Desnick RJ, Willner JP, Avraham KB, Ostrer H and others. A mutation of PCDH15 among Ashkenazi Jews with the type 1 Usher syndrome. N Engl J Med. 2003;348:1664–1670. doi: 10.1056/NEJMoa021502. [DOI] [PubMed] [Google Scholar]
- Broide E, Zeigler M, Eckstein J, Bach G. Screening for carriers of Tay-Sachs disease in the ultraorthodox Ashkenazi Jewish community in Israel. Am J Med Genet. 1993;47:213–215. doi: 10.1002/ajmg.1320470214. [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Ben-Yosef T, Dagan O, Frydman M, Abeliovich D, Sagi M, Abraham FA, Taitelbaum-Swead R, Shohat M, Hildesheimer M and others. The R245X mutation of PCDH15 in Ashkenazi Jewish children diagnosed with nonsyndromic hearing loss foreshadows retinitis pigmentosa. Pediatr Res. 2004;55:995–1000. doi: 10.1203/01.PDR.0000125258.58267.56. [DOI] [PubMed] [Google Scholar]
- Caggana M, Eng CM, Desnick RJ, Schuchman EH. Molecular population studies of Niemann-Pick disease Type A. Am J Hum Genet. 1994;55 Supplement:A147. [Google Scholar]
- Cao A, Rosatelli MC, Monni G, Galanello R. Screening for thalassemia: a model of success. Obstet Gynecol Clin North Am. 2002;29:305–328. doi: 10.1016/s0889-8545(01)00006-7. vi–vii. [DOI] [PubMed] [Google Scholar]
- Cohn JA, Neoptolemos JP, Feng J, Yan J, Jiang Z, Greenhalf W, McFaul C, Mountford R, Sommer SS. Increased risk of idiopathic chronic pancreatitis in cystic fibrosis carriers. Hum Mutat. 2005;26:303–307. doi: 10.1002/humu.20232. [DOI] [PubMed] [Google Scholar]
- DeMarchi JM, Caskey CT, Richards CS. Population-specific screening by mutation analysis for diseases frequent in Ashkenazi Jews. Hum Mutat. 1996;8:116–125. doi: 10.1002/(SICI)1098-1004(1996)8:2<116::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Dong J, Edelmann L, Bajwa AM, Kornreich R, Desnick RJ. Familial dysautonomia: detection of the IKBKAP IVS20(+6T --> C) and R696P mutations and frequencies among Ashkenazi Jews. Am J Med Genet. 2002;110:253–257. doi: 10.1002/ajmg.10450. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Dong J, Desnick RJ, Kornreich R. Carrier screening for mucolipidosis type IV in the American Ashkenazi Jewish population. Am J Hum Genet. 2002;70:1023–1027. doi: 10.1086/339519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Hashmi G, Song Y, Han Y, Kornreich R, Desnick RJ. Cystic fibrosis carrier screening: validation of a novel method using BeadChip technology. Genet Med. 2004;6:431–438. doi: 10.1097/01.gim.0000140836.66050.88. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Wasserstein MP, Kornreich R, Sansaricq C, Snyderman SE, Diaz GA. Maple syrup urine disease: identification and carrier-frequency determination of a novel founder mutation in the Ashkenazi Jewish population. Am J Hum Genet. 2001;69:863–868. doi: 10.1086/323677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstein J, Katzenstein H. The Dor Yeshorim story: community-based carrier screening for Tay-Sachs disease. Adv Genet. 2001;44:297–310. doi: 10.1016/s0065-2660(01)44087-9. [DOI] [PubMed] [Google Scholar]
- Ekstein J, Rubin BY, Anderson SL, Weinstein DA, Bach G, Abeliovich D, Webb M, Risch N. Mutation frequencies for glycogen storage disease Ia in the Ashkenazi Jewish population. Am J Med Genet A. 2004;129:162–164. doi: 10.1002/ajmg.a.30232. [DOI] [PubMed] [Google Scholar]
- Elpeleg ON, Anikster Y, Barash V, Branski D, Shaag A. The frequency of the C854 mutation in the aspartoacylase gene in Ashkenazi Jews in Israel. Am J Hum Genet. 1994;55:287–288. [PMC free article] [PubMed] [Google Scholar]
- Eng CM, Desnick RJ. Experiences in molecular-based prenatal screening for Ashkenazi Jewish genetic diseases. Adv Genet. 2001;44:275–296. doi: 10.1016/s0065-2660(01)44086-7. [DOI] [PubMed] [Google Scholar]
- Eng CM, Schechter C, Robinowitz J, Fulop G, Burgert T, Levy B, Zinberg R, Desnick RJ. Prenatal genetic carrier testing using triple disease screening. JAMA. 1997;278:1268–1272. [PubMed] [Google Scholar]
- Falik-Zaccai TC, Kfir N, Frenkel P, Cohen C, Tanus M, Mandel H, Shihab S, Morkos S, Aaref S, Summar ML and others. Population screening in a Druze community: the challenge and the reward. Genet Med. 2008;10:903–909. doi: 10.1097/GIM.0b013e31818d0e0f. [DOI] [PubMed] [Google Scholar]
- Fares F, Badarneh K, Abosaleh M, Harari-Shaham A, Diukman R, David M. Carrier frequency of autosomal-recessive disorders in the Ashkenazi Jewish population: should the rationale for mutation choice for screening be reevaluated? Prenat Diagn. 2008;28:236–241. doi: 10.1002/pd.1943. [DOI] [PubMed] [Google Scholar]
- Feigenbaum A, Moore R, Clarke J, Hewson S, Chitayat D, Ray PN, Stockley TL. Canavan disease: carrier-frequency determination in the Ashkenazi Jewish population and development of a novel molecular diagnostic assay. Am J Med Genet A. 2004;124:142–147. doi: 10.1002/ajmg.a.20334. [DOI] [PubMed] [Google Scholar]
- Grody WW, Cutting GR, Klinger KW, Richards CS, Watson MS, Desnick RJ. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–154. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- Gross SJ, Pletcher BA, Monaghan KG. Carrier screening in individuals of Ashkenazi Jewish descent. Genet Med. 2008;10:54–56. doi: 10.1097/GIM.0b013e31815f247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YS, Korman SH, Lee J, Ghoshal P, Wu Q, Barash V, Kang S, Oh S, Kwon M, Gutman A and others. Identification of a common mutation (Gly194Cys) in both Arab Moslem and Ashkenazi Jewish patients with dihydrolipoamide dehydrogenase (E3) deficiency: possible beneficial effect of vitamin therapy. J Inherit Metab Dis. 2003;26:816–818. doi: 10.1023/b:boli.0000010004.12053.5b. [DOI] [PubMed] [Google Scholar]
- Horowitz M, Pasmanik-Chor M, Borochowitz Z, Falik-Zaccai T, Heldmann K, Carmi R, Parvari R, Beit-Or H, Goldman B, Peleg L and others. Prevalence of glucocerebrosidase mutations in the Israeli Ashkenazi Jewish population. Hum Mutat. 1998;12:240–244. doi: 10.1002/(SICI)1098-1004(1998)12:4<240::AID-HUMU4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kaback M, Lim-Steele J, Dabholkar D, Brown D, Levy N, Zeiger K. Tay-Sachs disease--carrier screening, prenatal diagnosis, and the molecular era. An international perspective, 1970 to 1993. The International TSD Data Collection Network. JAMA. 1993;270:2307–2315. [PubMed] [Google Scholar]
- Kaback MM. Population-based genetic screening for reproductive counseling: the Tay-Sachs disease model. Eur J Pediatr. 2000;159 Suppl 3:S192–S195. doi: 10.1007/pl00014401. [DOI] [PubMed] [Google Scholar]
- Kalman L, Wilson JA, Buller A, Dixon J, Edelmann L, Geller L, Highsmith WE, Holtegaard L, Kornreich R, Rohlfs EM and others. Development of genomic DNA reference materials for genetic testing of disorders common in people of ashkenazi jewish descent. J Mol Diagn. 2009;11:530–536. doi: 10.2353/jmoldx.2009.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornreich R, Ekstein J, Edelmann L, Desnick RJ. Premarital and prenatal screening for cystic fibrosis: experience in the Ashkenazi Jewish population. Genet Med. 2004;6:415–420. doi: 10.1097/01.gim.0000139510.00644.f7. [DOI] [PubMed] [Google Scholar]
- Lebet T, Chiles R, Hsu AP, Mansfield ES, Warrington JA, Puck JM. Mutations causing severe combined immunodeficiency: detection with a custom resequencing microarray. Genet Med. 2008;10:575–585. doi: 10.1097/gim.0b013e31818063bc. [DOI] [PubMed] [Google Scholar]
- Lehavi O, Aizenstein O, Bercovich D, Pavzner D, Shomrat R, Orr-Urtreger A, Yaron Y. Screening for familial dysautonomia in Israel: evidence for higher carrier rate among Polish Ashkenazi Jews. Genet Test. 2003;7:139–142. doi: 10.1089/109065703322146830. [DOI] [PubMed] [Google Scholar]
- Lerner BH. When diseases disappear--the case of familial dysautonomia. N Engl J Med. 2009;361:1622–1625. doi: 10.1056/NEJMp0809587. [DOI] [PubMed] [Google Scholar]
- Li L, Eng C, Desnick RJ, German J, Ellis NA. Carrier frequency of the Bloom syndrome blmAsh mutation in the Ashkenazi Jewish population. Mol Genet Metab. 1998;64:286–290. doi: 10.1006/mgme.1998.2733. [DOI] [PubMed] [Google Scholar]
- Monaghan KG, Feldman GL, Palomaki GE, Spector EB. Technical standards and guidelines for reproductive screening in the Ashkenazi Jewish population. Genet Med. 2008;10:57–72. doi: 10.1097/GIM.0b013e31815f6eac. [DOI] [PubMed] [Google Scholar]
- Ness SL, Ben-Yosef T, Bar-Lev A, Madeo AC, Brewer CC, Avraham KB, Kornreich R, Desnick RJ, Willner JP, Friedman TB and others. Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet. 2003;40:767–772. doi: 10.1136/jmg.40.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorowicz A, Wilson BA, Schoor KP, Inoue H, Glaser B, Landau H, Stanley CA, Thornton PS, Clement JPt, Bryan J and others. Mutations in the sulonylurea receptor gene are associated with familial hyperinsulinism in Ashkenazi Jews. Hum Mol Genet. 1996;5:1813–1822. doi: 10.1093/hmg/5.11.1813. [DOI] [PubMed] [Google Scholar]
- Patrinos GP, Kollia P, Papadakis MN. Molecular diagnosis of inherited disorders: lessons from hemoglobinopathies. Hum Mutat. 2005;26:399–412. doi: 10.1002/humu.20225. [DOI] [PubMed] [Google Scholar]
- Peleg L, Pesso R, Goldman B, Dotan K, Omer M, Friedman E, Berkenstadt M, Reznik-Wolf H, Barkai G. Bloom syndrome and Fanconi's anemia: rate and ethnic origin of mutation carriers in Israel. Isr Med Assoc J. 2002;4:95–97. [PubMed] [Google Scholar]
- Petit C. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2:271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- Sansaricq C, Pardo S, Balwani M, Grace M, Raymond K. Biochemical and molecular diagnosis of lipoamide dehydrogenase deficiency in a North American Ashkenazi Jewish family. J Inherit Metab Dis. 2006;29:203–204. doi: 10.1007/s10545-006-0175-5. [DOI] [PubMed] [Google Scholar]
- Schrijver I, Kulm M, Gardner PI, Pergament EP, Fiddler MB. Comprehensive arrayed primer extension array for the detection of 59 sequence variants in 15 conditions prevalent among the (Ashkenazi) Jewish population. J Mol Diagn. 2007;9:228–236. doi: 10.2353/jmoldx.2007.060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchman EH, Miranda SR. Niemann-Pick disease: mutation update, genotype/phenotype correlations, and prospects for genetic testing. Genet Test. 1997;1:13–19. doi: 10.1089/gte.1997.1.13. [DOI] [PubMed] [Google Scholar]
- Shaag A, Saada A, Berger I, Mandel H, Joseph A, Feigenbaum A, Elpeleg ON. Molecular basis of lipoamide dehydrogenase deficiency in Ashkenazi Jews. Am J Med Genet. 1999;82:177–182. [PubMed] [Google Scholar]
- Shahrabani-Gargir L, Shomrat R, Yaron Y, Orr-Urtreger A, Groden J, Legum C. High frequency of a common Bloom syndrome Ashkenazi mutation among Jews of Polish origin. Genet Test. 1998;2:293–296. doi: 10.1089/gte.1998.2.293. [DOI] [PubMed] [Google Scholar]
- Strom CM, Crossley B, Redman JB, Quan F, Buller A, McGinniss MJ, Sun W. Molecular screening for diseases frequent in Ashkenazi Jews: lessons learned from more than 100,000 tests performed in a commercial laboratory. Genet Med. 2004;6:145–152. doi: 10.1097/01.gim.0000127267.57526.d1. [DOI] [PubMed] [Google Scholar]
- Strom CM, Janeczko RA, Anderson B, Redman J, Quan F, Buller A, McGinniss MJ, Sun WM. Technical validation of a multiplex platform to detect thirty mutations in eight genetic diseases prevalent in individuals of Ashkenazi Jewish descent. Genet Med. 2005;7:633–639. doi: 10.1097/01.gim.0000187120.93597.16. [DOI] [PubMed] [Google Scholar]
- Strom CM, Janeszco R, Quan F, Wang SB, Buller A, McGinniss M, Sun W. Technical validation of a TM Biosciences Luminex-based multiplex assay for detecting the American College of Medical Genetics recommended cystic fibrosis mutation panel. J Mol Diagn. 2006;8:371–375. doi: 10.2353/jmoldx.2006.050115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Grody WW. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. J Mol Diagn. 2008;10:484–492. doi: 10.2353/jmoldx.2008.080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlander PC, Kaporis A, Liu Q, Zhang Q, Seligsohn U, Auerbach AD. Carrier frequency of the IVS4 + 4 A-->T mutation of the Fanconi anemia gene FAC in the Ashkenazi Jewish population. Blood. 1995;86:4034–4038. [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Waldmuller S, Muller M, Rackebrandt K, Binner P, Poths S, Bonin M, Scheffold T. Array-based resequencing assay for mutations causing hypertrophic cardiomyopathy. Clin Chem. 2008;54:682–687. doi: 10.1373/clinchem.2007.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson MS, Cutting GR, Desnick RJ, Driscoll DA, Klinger K, Mennuti M, Palomaki GE, Popovich BW, Pratt VM, Rohlfs EM and others. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman S, Lahad A, Shmueli A, Zimran A, Peleg L, Orr-Urtreger A, Levy-Lahad E, Sagi M. Carrier screening for Gaucher disease: lessons for low-penetrance, treatable diseases. JAMA. 2007;298:1281–1290. doi: 10.1001/jama.298.11.1281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.