Abstract
Rare copy number variations (CNVs) are a recognized cause of common human disease. Predicting the genetic element(s) within a small CNV whose copy number loss or gain underlies a specific phenotype might be achieved reasonably rapidly for single patients. Identifying the biological processes that are commonly disrupted within a large patient cohort which possess larger CNVs, however, requires a more objective approach that exploits genomic resources. In this study, we first identified 98 large, rare CNVs within patients exhibiting multiple congenital anomalies. All patients presented with global developmental delay (DD), while other secondary symptoms such as cardiac defects, craniofacial features and seizures were varyingly presented. By applying a robust statistical procedure that matches patients’ clinical phenotypes to laboratory mouse gene knockouts, we were able to strongly implicate anomalies in brain morphology and, separately, in long-term potentiation as manifestations of these DD patients’ disorders. These and other significantly enriched model phenotypes provide insights into the pathoetiology of human DD and behavioral and anatomical secondary symptoms that are specific to DD patients. These enrichments set apart 103 genes, from among thousands overlapped by these CNVs, as strong candidates whose copy number change causally underlies approximately 46% of the cohort's DD syndromes and between 59 and 80% of the cohort's secondary symptoms. We also identified significantly enriched model phenotypes among genes overlapped by CNVs in both DD and learning disability cohorts, indicating a congruent etiology. These results demonstrate the high predictive potential of model organism phenotypes when implicating candidate genes for rare genomic disorders.
INTRODUCTION
A rapidly increasing proportion of human genetic diseases are thought to arise from copy number variations (CNVs), defined as >1 kb duplicated or deleted stretches of DNA (1). Phenotypically, some of these disorders manifest as multiple congenital anomalies, which generally include developmental delays (DDs) along with variable secondary features such as cardiac defects and cranio-facial differences (2–5). Children with DD fail to achieve normal developmental milestones, both physical and intellectual, in early childhood (<5 years), and often have impaired motor function, cognitive ability and/or language skills. Delayed or impaired neurological development frequently leads to learning disabilities (LD; also termed mental retardation).
In the majority of DD cases, the identities of causative genes, however, remain unknown, particularly for large CNVs encompassing many genes. In individual cases, a clinical geneticist may highlight an excellent candidate gene for DD on the basis of prior experience and by sampling the available literature. This process, however, is inevitably subjective and time-consuming, and it necessarily rests on the completeness, availability and easy accessibility of a rapidly increasing corpus of knowledge. Such a process will also fail to discover molecular pathways or processes whose disruption has not been reported previously as being associated with DD. Accurate definition of disease-relevant pathways or processes, however, remains far from straightforward, as the available electronic pathway resources, including the Gene Ontology (6) (GO) and Kyoto Encyclopedia of Genes and Genomes (7) (KEGG) do not capture the true complexity of disease-relevant biological pathways or processes. The identification of DD-relevant genes is further complicated by the presence of large numbers of CNVs in the general, apparently healthy, population (8–10). If, however, it is assumed that such variants do not contribute to the pathoetiology of developmental conditions, then their genes can be excluded when seeking disease-relevant genes.
Our goal in this study was to obtain evidence, using a robust statistical approach, for the causative element(s) underlying each patient's clinical presentation. More specifically, we sought to identify disruptive genetic changes among a large cohort of 87 individuals, providing statistical genetic evidence not only for their DD presentations, but also for their additional phenotypes, such as behavior or eye abnormalities. To identify genes and biological processes that underlie these patients’ phenotypes, we turned to an experimental resource which is orthogonal to, and likely more relevant than, electronic molecular pathways. This is a set of 5329 defined phenotypes associated with 5011 genes disrupted in mouse models that have been organized in a phenotype ontology (11). We hypothesized that each disease-causative CNV region will harbor one or more gene(s) whose mouse ortholog, when disrupted, results in a phenotype that corresponds to that of the human disease under investigation. Furthermore, if sufficient CNV regions with similar disease associations were to be known, particularly those containing relatively few genes, then it might be possible to detect within these regions significant enrichments of genes whose orthologs, when disrupted, result in particular mouse phenotypes that are relevant to that disease. This approach seeks significant associations between patients’ genotypes and what we term ‘model phenotypes’ observed for the knockout models of orthologous mouse genes. Together with their associated mouse knockouts, these model phenotypes are available to provide useful insights into the molecular and cellular pathoetiology of disease. If this strategy is to be successful, then it must control the rate of false discovery associations that inevitably accrue from the large number of statistical tests―one for each phenotype―that are being applied.
In a previous study, we applied a similar but more primitive approach to 148 de novo CNV intervals from LD individuals (12). Among over 200 diverse nervous system phenotypes that were investigated, we identified two mouse model phenotypes that were significantly over-represented with a low false discovery rate (FDR) <5%. Each of these model phenotypes, abnormal axon morphology and abnormal dopaminergic neuron morphology, is of particular relevance to human LD phenotypes. We were also able to demonstrate significant associations between human and model phenotypes for additional clinical features other than LD that were apparent from this patient population (12).
We considered it important to develop our novel methodology further and to apply it more widely to determine whether it provides insights into other patient datasets. We sought to investigate whether the method is effective in highlighting candidate genes and biological processes in seemingly heterogeneous syndromes, which present the greatest challenges for this, and other functional enrichment, approaches. Thus, we wished to know (i) whether the candidate causative elements identified among these large, multigenic CNVs would indicate a single biological process as explaining a shared DD phenotype, or else would stratify the cohort on the basis of different biological processes which would be suggestive of the disorder being heterogeneous in etiology; (ii) whether multiple elements within each patient's CNV(s) contribute additively or multiplicatively to the disorder; (iii) whether functional elements contribute to both primary and secondary features of a patient or whether pleiotropy is indicated; (iv) whether there is a qualitative or a quantitative difference among the functional elements identified for either Gain or Loss CNVs; and finally, (v) whether DD and LD are associated with comparable model phenotypes.
By applying an extended mouse phenotype method to a set of 98 CNVs observed in individuals with DD, we identified significant associations of model phenotypes with CNV genes which subsequently allowed a set of commonly disrupted biological processes to be identified and 103 candidate genes to be collated. These represent excellent candidates for genes whose copy number change contribute to DD and associated phenotypes. Model phenotypes thus provide valuable insights into the pathoetiology of DD for single individuals, and pinpoint genes whose copy number change likely underlies their, and other DD patients’, phenotypes.
RESULTS
The genomes of 87 patients, each of whom has been clinically diagnosed with diverse DD syndromes, were examined using high-density oligonucleotide microarrays that allowed the identification of CNVs that might be causative of their disorders. Ninety-eight de novo CNVs were identified in 87 individuals (Supplementary Material, Table S1). In addition, these CNVs were not detected in multiple CNV databases generated from large numbers of healthy control individuals (see Materials and Methods). Their absence from parent and control samples suggests that these CNVs have arisen spontaneously in each patient. Patients’ CNVs ranged in size from 10.5 kb to 56.1 Mb (median 5.3 Mb, total 609.1 Mb) and completely overlap 3834 protein-coding genes that are also not overlapped by CNVs observed in apparently healthy individuals (Table 1; see Materials and Methods). The observation that these CNVs tend to be over an order of magnitude larger than CNVs detected in the genomes of apparently healthy individuals (Table 1) accords well with their likely pathogenicity.
Table 1.
Genomic extent and NCBI gene content for DD-associated CNVs and benign CNVs. The genes considered are those remaining after excluding genes also overlapped by benign CNVs in the same direction of copy change (see Materials and Methods)
| CNV number (median size) | CNVR number (median size) | Gene count | Genes with mouse KO orthologs | Genome covered (Mb) | |
|---|---|---|---|---|---|
| Benign | 26,472 (0.21 Mb) | 1388 (0.17 Mb) | 4576 | 681 | 429.0 |
| DD All | 98 (4.85 Mb) | 65 (5.34 Mb) | 3834 | 808 | 609.1 |
| DD Gain | 30 (4.38 Mb) | 24 (4.73 Mb) | 1908 | 401 | 283.3 |
| DD Loss | 68 (5.30 Mb) | 51 (5.35 Mb) | 2263 | 468 | 374.3 |
Linking DD CNV genes to model phenotypes
Identifying individual genes whose copy number change has substantially contributed to heterogeneous DD phenotypes is greatly hindered by the large numbers of genes (median 40.5, mean 60.6) located within these DD-associated CNVs. To identify one or few candidates, among these dozens of genes, that are likely to contribute to DD phenotypes we adopted a two-stage strategy. First, by applying rigorous statistical tests, we sought evidence that specific classes of genes (those with particular phenotype annotations) are significantly over-represented within these DD-associated copy number variation regions (CNVRs). We then derived a set of candidate genes drawn from all loci that are: (i) present within these CNVRs, and (ii) annotated with at least one of these over-represented classes.
To identify classes of genes enriched in these DD-associated CNVs, we investigated whether they randomly sample genes that, when disrupted in mice, result in specific nervous system phenotypes. We chose to consider only nervous system phenotypes because of their obvious relevance to DD patients who all exhibit neurological deficits. We first assembled 98 DD-associated CNVs into 65 distinct CNVRs (Table 1). Several overlapping CNVs were observed in opposite directions of copy number change. To investigate whether the direction of copy change might reveal distinct pathoetiologies with equally distinct genetic causes, we divided by direction of change and separately assembled 24 Gain CNVRs and 51 Loss CNVRs (Table 1). For each set of Gain, Loss or All CNVRs, we considered only those genes that were overlapped by their CNVRs and that were not overlapped by ‘benign’ CNVs in apparently healthy control individuals (see Materials and Methods). For each set, we then examined whether the mouse orthologs of these genes were significantly enriched in any of 147 nervous system phenotypic terms after controlling for false-discovery associations (FDR <5%).
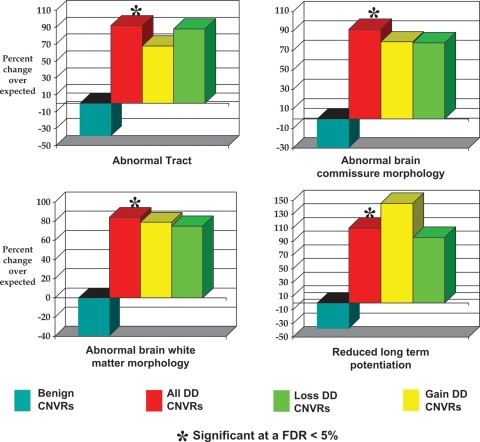
Four model phenotypes were identified that were each substantially and significantly enriched among All DD-associated CNVRs (Table 2 and Fig. 1). The first three of these phenotypes (abnormal tract, abnormal brain white matter morphology and abnormal brain commissure morphology) are closely related within the hierarchy of mouse phenotypes. The ∼2-fold increase in the numbers of genes in these CNVRs whose orthologs, when disrupted, present these anatomical anomalies strongly implicates them as being a distinguishing feature of these DD patients’ genotypes compared with the general population. Twenty-six genes contribute to the significant associations between DD-associated CNVRs and all three model phenotypes, while a further six genes contribute only to either abnormal tract and/or abnormal brain white matter morphology terms (Table 3).
Table 2.
Four mouse knockout phenotypes that are significantly enriched for the set of All DD-associated CNVRs (FDR <5%)
| MGI phenotype | Mammalian phenotype accession | Total human genes with a mouse ortholog yielding this phenotype | Definition | Observed | Expected | Enrichment | P-value |
|---|---|---|---|---|---|---|---|
| Abnormal tract | MP:0000778 | 97 | Anomaly in the structure of any bundle of myelinated nerve fibers following a defined path through the brain and/or spinal cord | 30 | 15.6 | +92% | 1.8 × 10−4 |
| Abnormal brain white matter morphology | MP:0008026 | 98 | Any structural anomaly of the regions of the brain that are largely or entirely composed of myelinated nerve cell axons and contain few or no neural cell bodies or dendrites | 29 | 15.8 | +84% | 5.3 × 10−4 |
| Abnormal brain commissure morphology | MP:0002199 | 84 | Any structural anomaly of any of the nerve fiber tracts that span the longitudinal fissure between the cerebral and/or cerebellar hemispheres of the brain | 26 | 13.5 | +92% | 4.8 × 10−4 |
| Reduced long-term potentiation | MP:0001473 | 71 | Less than the normal persistent robust synaptic response induced by synchronous stimulation of pre- and post-synaptic cells | 24 | 11.4 | +110% | 1.8 × 10−4 |
Figure 1.
Enrichments of MGI phenotype terms among genes overlapped by DD-associated CNVRs. Four specific nervous system phenotypes (abnormal tract, abnormal brain commissure morphology, abnormal brain white matter morphology and reduced long-term potentiation) are significantly over-represented in genes overlapped by All CNVRs. The phenotypes result from the disruption of mouse genes that have been mapped to their unique human ortholog. Columns marked with an asterisk (‘*’) are significantly enriched (FDR <5%).
Table 3.
Candidate genes for DD and associated clinical features. These are present in DD-associated CNVRs and belong to any of four significantly enriched annotations; namely, mouse knockout phenotypes of abnormal tract, abnormal brain white matter morphology, abnormal brain commissure morphology and reduced long-term potentiation genes (Fig. 1). The remaining genes lie within CNVs associated with the particular secondary clinical features and belong to significant enrichments identified as specific to those clinical feature (see main text and Figs 2–4)
| MGI phenotype | Gene in Loss DD CNVR |
Gene in Gain DD CNVR |
||||
|---|---|---|---|---|---|---|
| Associated with DD | ||||||
| Abnormal tract (MP:0000778) | AKT3, APP, CELSR3, DCC, DCLK1, EFNB3 | ENAH, EPHA8, EPHB2, FGF2, HTT, MARCH7 | MYCBP2, NFIA, PSEN2a, SEMA3F, SIM1 | CLIP2, EFNB2, EPHA4, EPHB1 | EXT1, HSF1, LYNX1, MAP2b, MAPK8IP3 | NCK1c, NFIB, OTX1d, PTK2e, ST8SIA2f |
| Abnormal brain white matter morphology (MP:0008026) | AKT3, APP, ARSA, CELSR3, DCC, DCLK1 | ENAH, EPHB2, FGF2, HTT, MARCH7, MYCBP2 | NFIA, PSEN2, QKI, SEMA3F | CLIP2, EFNB2, EPHA4, EPHB1, EXT1 | HSF1, LYNX1, MAP2, NCK1 | NFIB, OTX1, PTK2, QKI, ST8SIA2 |
| Abnormal brain commissure morphology (MP:0002199) | AKT3, APP, CELSR3, DCC, DCLK1 | ENAH, EPHB2, FGF2, HTT, MARCH7 | MYCBP2, NFIA, PSEN2a, SEMA3F | CLIP2, EFNB2, EPHA4, EXT1 | HSF1, LYNX1, MAP2b, NCK1c | NFIB, OTX1d, PTK2e, ST8SIA2f |
| Reduced long-term potentiation (MP:0001473) | APP, B3GAT1, DOC2A, EFNB3, GRIK2 | GRM5, JPH3, LEPR, MAPK3 | OPRM1, RIMS1, TNC, VLDLR | ADCY8, ADD2, ARC, EFNB2, EPHA4 | JPH3, NCAM1, OPRM1, PRKAR1B, SERPINE2 | SLC24A2, STX1A, THY1, TNC |
| Associated with behavioral abnormality | ||||||
| Abnormal cued conditioning behavior (MP) | APP, GRIK2 | MAPK1 | PRKCB1 | ARC, CLIP2, LIMK1 | LYNX1, MAPK1 | PRKCB1, TNC |
| Abnormal conditioning behavior (MP) | APP, DOC2A, GRIK2 | MAPK1, MAPK3 | PRKCB1, TNC | ARC, CLIP2, LIMK1 | LYNX1, MAPK1 | STX1A TNC |
| Decreased fear-related response (MP) | APP | GRIK2 | TNC | ARC | TNC | |
| Abnormal brain white matter morphology (MP) | APP | QKI | CLIP2, EPHB1, HSF1 | LYNX1, NCK1c | PTK2e QKI | |
| Increased sensitivity to addictive substance | GNAZ | OPRM1 | GNAZ | LYNX1 | OPRM1 | |
| Associated with eye abnormalities | ||||||
| Abnormal ocular fundus morphology | ATP1B2, BCL2, DCC, EDNRB | FAM48A, GUCY2D, LMO7, MAB21L1 | RB1g, TP53, UCHL3 | ALDH1A1, RORB | SLC4A3 | XRCC5 |
| Abnormal retina morphology | ATP1B2, BCL2, DCC, FAM48A | GUCY2D, LMO7, MAB21L1 | RB1g, TP53, UCHL3 | ALDH1A1, RORB | SLC4A3 | XRCC5 |
| Abnormal retinal layer morphology | ATP1B2, BCL2, DCC | FAM48A, GUCY2D, LMO7 | RB1g, TP53, UCHL3 | RORB | ||
| Abnormal retinal apoptosis (MP) | RB1g | TP53 | UCHL3 | SLC4A3 | XRCC5 | |
| Associated with seizures | ||||||
| Abnormal voluntary movement (MP) | ARSA, DCC, ENAH, ESR1, F5, FGFR3, FGFRL1, FIGN, GNAL, GPR161 | HTT, IDUA, MAPK3, MC2R MC4R, MMP14, NOX3, OPRM1, PARK2, PDE10A | PSEN2a PTPN2, QKI, RGS7, SCN1A, SCN3A, SELEh, SELPh, SLC19A2, SLC7A8, Ti, TACC3 | EFNB2, ESR1, FGF12, FGF14, NOX3 | OPRM1, PARK2, PCCA, PDE10A, QKI | T, ZIC2, ZIC5 |
| Abnormal stationary movement (MP) | FIGN, GPR161, HTT | NOX3 PARK2, QKI, SCN1A | SELEh, SELPh, Ti, TACC3 | EFNB2, NOX3, PARK2 | PCCA, QKI | T, ZIC2 |
| Abnormal, emotion/affect, behavior (MP) | ESR1, GNAL, HTT | MC5R, OPRM1, PARK2 | PDE10A, QKI, RGS7 | ESR1, FGF12, MAS1 | OPA1, OPRM1, PARK2 | PDE10A, QKI |
| Abnormal response to novelty (MP) | GNAL, HTT | PARK2, PDE10A | QKI, RGS7 | FGF12, OPA1 | PARK2, PDE10A | QKI |
aPhenotype results from a dual Psen1 and Psen2 disruption.
bPhenotype results from a dual Mtap2 and Mtap1b disruption.
cPhenotype results from a dual Nck1 and Ncbk2 disruption.
dPhenotype results from a dual Otx1 and Otx2 disruption.
ePhenotype results from a dual Ptk2 and Emx1 disruption.
fPhenotype results from a dual ST8SIA2 and ST8SIA4 disruption.
gPhenotype results from a dual Rb1 and Rbl1 disruption.
hPhenotype results from a dual Sele and Selp disruption.
iPhenotype results from a dual T and Nox3 disruption.
The fourth phenotype (reduced long-term potentiation; rLTP; Table 2) appears to have been identified largely independently of the other phenotypes, as only four of the 24 contributing DD-associated CNV genes that possess the rLTP model phenotype also exhibit any of the three other model phenotypes (Table 3). The association between these patients’ CNVR genes and the rLTP model phenotype strongly implicates deficits in synaptic plasticity and long-term memory as contributing strongly to human DD phenotypes. It is important to note that, as expected, genes associated with each of these four model phenotypes are substantially depleted among the set of 4576 genes overlapped by apparent benign CNVs (Fig. 1).
Model phenotypes for secondary symptoms of DD individuals
The success in associating model phenotypes to, and identifying candidate genes for, the primary DD phenotype then prompted us to apply our statistical approach to these patients’ secondary phenotypes. For 74 of the 87 DD patients, there were clinical data available that indicated one or more secondary clinical phenotypes in addition to DD (Supplementary Material, Table S2). Twelve major secondary phenotype categories were defined in the cohort, namely behavioral abnormalities, brain malformations, cardiac defects, cleft lip, cleft palate, facial dysmorphia, eye abnormalities, sensorineural hearing loss, limb abnormalities, seizures, short stature and urogenital symptoms (further explained in Materials and Methods). To test for associations between these secondary phenotypes and CNV genes, we first grouped patients’ CNVs non-exclusively according to the 12 phenotypes and assembled them into CNVRs (see Materials and Methods). Next, we tested for enrichments of human genes in these CNVRs whose mouse orthologs, when disrupted, exhibit phenotypes drawn from only the most relevant categories of mouse phenotypes (Supplementary Material, Table S3). For example, genes in CNVs from patients exhibiting brain malformations were tested for association with nervous system, but not pigmentation, phenotypes. Categories that were tested each contained between 129 and 220 terms and, as before, we applied a stringent correction for multiple tests (FDR <5%).
Significant associations were identified for three secondary symptom classes for these patients, namely behavioral abnormalities, seizures and eye abnormalities (Figs 2–4, respectively). Importantly, and as expected, these enrichments are observed to be specific only to those CNVRs associated with the particular secondary symptom and not to CNVRs from patients not presenting with that particular secondary symptom (Figs 2–4). For example, patients not presenting with a behavioral phenotype were not associated with a behavioral phenotype in mouse knockout experiments (brown, black and tan bars in Fig. 2). The specificity of these enrichments to those patients presenting with a particular secondary symptom also implies that under-ascertainment of these secondary symptoms within the DD cohort is not apparent.
Figure 2.
Enrichments of MGI phenotype terms among genes overlapped by CNVRs from DD patients who exhibit behavioral abnormalities. Four specific behavior/neurological phenotypes (Abnormal cued conditioning behavior, Abnormal conditioning behavior, Increased sensitivity to addictive substance and Decreased fear related response) and one specific nervous system phenotype (Abnormal Brain White Matter Morphology) are significantly over-represented in genes overlapped by All and/or Gain behavioral abnormality-associated CNVRs. The phenotypes result from the disruption of mouse genes that have been mapped to their unique human ortholog. Columns marked with an asterisk (‘*’) are significantly enriched (FDR <5%).
Figure 4.
Enrichments of MGI phenotype terms among genes overlapped by CNVRs from DD patients who exhibit eye abnormalities. Although 11 eye phenotypes are found to be significantly over-represented in Loss eye abnormality-associated CNVRs, we show only the four principal phenotypes here (Abnormal ocular fundus morphology, Abnormal retina morphology, Abnormal retina apoptosis and Abnormal retinal layer morphology); all 11 significant eye-associated phenotypes are shown in Supplementary Material, Figure S1. The phenotypes result from the disruption of mouse genes that have been mapped to their unique human ortholog. Columns marked with an asterisk (‘*’) are significantly enriched (FDR <5%).
Figure 3.
Enrichments of MGI phenotype terms among genes overlapped by CNVRs from DD patients who exhibit seizures. Four specific behavior/neurological phenotypes (Abnormal voluntary movement, Abnormal stationary movement, Abnormal emotion/affect behavior and Abnormal response to novelty) are significantly over-represented in genes overlapped by Gain seizure-associated CNVRs. The phenotypes result from the disruption of mouse genes that have been mapped to their unique human ortholog. Columns marked with an asterisk (‘*’) are significantly enriched (FDR <5%).
Many of these model phenotype associations for secondary symptoms obtain significance among either Gain or Loss CNVRs but not among All CNVRs. Indeed, model phenotype associations with the patients’ seizures phenotype are found only among Gain CNVRs, while associations with eye abnormalities are observed only among Loss CNVRs (Figs 3 and 4). CNVRs in the opposite copy number change direction (i.e. Loss and Gain, respectively) appear little different from CNVRs not associated with these secondary symptoms. These Gain- or Loss-specific enrichments appear unlikely to have resulted from diminished power within particular CNV subsets (seizures-associated CNVs: 256 Gain genes, Loss 962 genes; eye symptoms-associated CNVs: 482 Loss genes, 501 Gain genes). The seizures model phenotype thus appears to be specifically associated with Gain CNVs, while eye model phenotypes are specifically associated with Loss CNVs.
Model phenotypes identify candidate genes
Our findings reflect the non-random concentration of genes with particular annotations (specifically, mouse model phenotypes) that overlap DD-associated CNVRs. Such genes thus become strong candidates for causative elements whose copy number change underlies the DD disorder for individual patients (Table 2 and Supplementary Material, Table S4). The four significantly enriched model phenotypes associated with the primary DD phenotype (abnormal brain white matter morphology, abnormal brain commissure morphology, abnormal tract and reduced long-term potentiation) provide 52 candidate genes that are overlapped by DD-associated CNVs (Table 3 and Supplementary Material, Table S4). By randomly sampling 1000 gene sets of equal number (see Materials and Methods), we find that this represents a large 86% increase over the number expected by chance (random samples’ distribution median 28, SD 5). In addition, we identify 72 genes associated with model phenotypes for secondary symptoms, of which 21 are also associated with the primary DD presentation. All but one (namely, DCC) of these 21 genes associated with both primary and secondary presentations represent neurological phenotypes. This is consistent with abnormal neural development often resulting in multiple developmental effects.
Candidate genes for DD are relevant to 42 of 98 (43%) DD-associated CNVs and 39 of 87 (45%) DD patients (Supplementary Material, Table S4). Among all CNVs harboring one or more DD-associated candidate gene, the median number of such genes per CNV is 1 (mean 1.6, SD 1.0), with a maximum of 6. However, Gain CNVs contain significantly more such genes than Loss CNVs (Mann–Whitney U test, P = 0.003; candidate genes per Gain CNV: median 2, mean 2.2, SD 1.4; candidate genes per Loss CNV: median 1, mean 1.3, SD 0.4; Supplementary Material, Table S4) despite Gain CNVs tending to be physically smaller than Loss CNVs (median 4.38 versus 5.40 Mb, respectively; Table 1). Although we must sound a note of caution resulting from the incomplete coverage of human genes by mouse knockout phenotypes (see Discussion), this result suggests that for DD to be revealed in the clinic, Gain DD-associated CNVs will tend to involve more causative genes than Loss DD-associated CNVs. This is expected if gene deletions are more deleterious than gene duplications.
The model phenotypes identified for seizures, behavior or eye abnormality secondary symptoms of DD patients yield candidate genes for 60–78% of associated CNVs and 59–80% of associated patients (Supplementary Material, Table S4). Single CNVs often contain more than one secondary symptom-associated candidate gene (seizure-associated CNVs: median 2 genes, mean 3.1, SD 2.0; behavior abnormality-associated CNVs: median 2 genes, mean 1.8, SD 0.9; and eye abnormality-associated CNVs: median 2.5 genes, mean 3.0, SD 1.4). For both seizures and eye-related secondary symptoms, there are significantly more candidate genes per CNV than for the primary DD phenotype (Mann–Whitney U tests, P< 0.002). This implies that for these secondary symptoms to be manifested, multiple dosage-sensitive genes may often need to be affected.
As 11 of 87 DD patients harbor two, as opposed to one, large rare CNVs, we then considered whether multiple CNVs within an individual might contribute to the same enrichment. For patients presenting with seizures in this study this is indeed the case: the median number of seizure-associated candidate genes is 3.5 per patient versus 2 per CNV. These patients’ susceptibility to suffer seizures may thus result from gene copy number changes at more than one genomic location.
Comparison of DD- and LD-associated CNVs
DD is defined as the significant delay in reaching early developmental (both physical and mental) milestones, and thus often is considered to be etiologically similar to LD. Indeed, DD children are frequently diagnosed with LD at a later age. Consequently, we were interested in comparing the candidate genes and enriched model phenotypes identified for the DD cohort with those we identified previously for a separate LD cohort (12). Neither of the two model phenotypes found from 148 LD-associated CNVs was found to be significantly enriched in the DD-associated CNV gene set (data not shown). This may be interpreted as implying that the etiologies of DD and LD are divergent, or more likely that both disorders are etiologically heterogeneous resulting in findings not being replicated for different patient cohorts of small-to-medium size. Nevertheless, among the four model phenotypes identified here from DD-associated CNVs, genes whose orthologs are associated with abnormal tract are also significantly over-represented among genes overlapped by LD CNVs (+49% enrichment, P= 0.04; single test). For LD-associated CNVs, the abnormal tract enrichment segregates strongly with Loss CNVs (+73% enrichment, P= 0.009) where the enrichment is very similar to that observed within the DD-associated CNV genes (Fig. 1). Furthermore, among LD-associated Loss CNVs, the two related mouse model phenotypes for DD (Fig. 1) are also significantly enriched: abnormal brain white matter morphology (+70.8% enrichment, P= 0.01) and abnormal brain commissure morphology (+59.4%, P = 0.04). Consequently, there is evidence that DD and LD share a congruent etiology. Of 21 and 30 candidate genes identified from these two model phenotypes that are associated with LD and DD, respectively, five are seen in both patient cohorts, namely AKT3, HTT, SIM1, CLIP2 and SEMA3F. Variants of these five genes thus may contribute to both DD and LD disorders, thereby identifying a common molecular etiology for patients in both cohorts.
DISCUSSION
Our findings, and those from other studies, imply that cognitive disorders are highly heterogeneous in etiology. Indeed, most genes have been associated with these disorders only once. Thus, we might expect combinations of rare variants among several hundred genes to contribute to cognitive disorders (13). The allelic heterogeneity of these disorders compels us not to identify individual genes, but rather to identify those biological processes or pathways whose disruption results in developmental disease. A priori, it is unclear which genomic resource best predicts these processes or pathways, whether, for example, a specific molecular function (e.g. transcription factor activity) is more likely to explain these disorders than a more over-arching cellular function (e.g. regulation of cell growth).
Our results illustrate the utility and power of a complementary genomic resource, namely mouse phenotypic data, to identify strong candidate genes for developmental disorders from statistically robust enrichments. As mouse phenotype information is currently available for a minority (∼25%) of human genes, only the strongest signals are likely to be discovered and undoubtedly many relevant genes that contribute to complex and diverse phenotypes remain to be identified. Nonetheless, we have shown how four phenotypes identified in this study (rLTP, abnormal tract, abnormal white matter morphology and abnormal brain commissure morphology), together with their associated genes, provide promising lines of investigation into the contributions of CNVs and their genes to DD phenotypes. Furthermore, significant enrichments that are specifically associated with CNVs from patients presenting behavioral, eye or seizures secondary symptoms also are informative of disease etiology. For primary (DD) and secondary symptoms, we have identified 103 candidate genes across 56 (57%) CNVs derived from 50 (57%) DD patients (Table 2 and Supplementary Material, Table S4). Several of these candidate genes have previously been implicated in neurological disease, for example, NFIA (14), SCN1A (15), GRIK2 (16), VLDLR (17), ZIC2 (18) and FGF14 (19) (see Supplementary Material, Table S4 for additional OMIM annotations).
Concordance between model and human phenotypes
Significant association between DD CNV genes and the brain commissure model phenotype is consistent with previous observations of brain commissure abnormalities in patients with neurodevelopmental disorders (20). The mouse phenotype Abnormal brain commissure morphology covers several commissure abnormalities often seen in DD patients, involving the corpus callosum (21,22) (CC), the anterior commissure (22,23) and the hippocampal commissure (22,24). Indeed, CC abnormalities have frequently been associated with chromosomal aberrations such as CNVs (22,25). Furthermore, many genes whose mutations give rise to an abnormal CC have been observed to be haploinsufficient, with the severity of the abnormality often correlating with the reduction in copy number (20). Commissure abnormalities have been previously implicated in 3–5% of patients with neurodevelopmental disorders (20), while we observed that 25% (22/87) of DD patients have CNVs that overlap commissure-associated genes. This suggests that commissure abnormalities might be substantially more prevalent among DD patients than is readily apparent from non-invasive imaging, particularly for those often more subtle CC abnormalities arising from heterozygous mutations (20). Furthermore, many CC abnormality-associated genes appear to give variable phenotypic manifestations depending on the presence of other genetic modifiers (20,26,27), of which there are potentially many in these large DD CNVs.
We were also encouraged by the significant association of DD CNV genes with an rLTP model phenotype, as this result accords with previous descriptions of human DD phenotypes (28). More specifically, the roles of LTP in synaptic plasticity and in neural development have long been recognized (29). Reduced LTP, for example, is a prominent feature of mice that have been engineered to carry copy number gains that model mental retardation in Down Syndrome (30). In this respect, it may be relevant that the rLTP signal we identify in DD-associated CNVs is most enriched among Gain CNVs (Fig. 1).
The candidate genes identified for DD in this study include two transcription factors from the nuclear factor 1 gene family, NFIA and NFIB. The NFI gene family controls several important processes in CNS development including axon guidance, glial and neuronal cell differentiation, and neuronal migration (31). Furthermore, since the NFI gene family are transcription factors, they regulate the expression of several other neuronal and glial genes. Interestingly, expression of another DD candidate gene, TNC, is regulated by NFI transcription factors, as is EPHB1 (32,33), one of seven members of the Ephrin families of receptor protein-tyrosine kinases and their ligands involved in synapse formation and plasticity in the central nervous system (34) all identified as DD candidate genes in this study (EFNB2, EFNB3, EPHA4, EPHA8, EPHB1 and EPHB2). Similarly, multiple members of other gene families, like fibroblast growth factors and their receptors (namely FGF2, FGF12, FGF14, FGFR3 and FGFRL1), glutamate receptors (GRM5 and GRIK2) and members of the mitogen-activated protein kinase family (MAPK1 and MAPK3) were also identified as candidate genes for related phenotypes (Table 3). These observations indicate functional convergence and common signaling pathways underlying the neurological phenotypes observed in DD patients. Furthermore, several candidate genes are known to have functions relevant to DD and associated phenotypes, such as MAPK1 and MAPK3, both of which belong to the ERK MAP kinase signaling cascade which contributes to brain development, learning, memory and cognition (35). Among all these gene families with members implicated in causing DD in our study, only the Ephrin family has multiple candidate genes overlapped by the same CNV (EPHA8 and EPHB2, Supplementary Material, Table S4).
For all but seven of the 103 candidate genes (namely, NFIA, SCN1A, OPA1, OPRM1, DCLK1, MMP14 and DCC), the phenotype that contributes to the enrichments detected here describes the homozygous disruption of their mouse ortholog. Given that the DD-associated CNVs reported here are all apparently heterozygous, relating the homozygous loss in mouse to the hemizygous loss in human for the remaining 96 candidate genes might appear, at first, to be a difficulty. However, for all 32 (29%) of these genes for which hemizygous knockout phenotypes have been reported, none exhibit normal phenotypes and thus all 32 candidate genes can be considered as being haploinsufficient. Neither can it be assumed that where a hemizygous mouse phenotype has not been reported, the hemizygous state gives no phenotype. For example, whereas only the homozygous mouse knockout of Fgf14 is reported, hemizygous disruption of FGF14 in humans is demonstrably pathogenic (19,36).
Forty-three percent of DD patients whose CNVs we have been able to suggest candidate genes show a copy number change in more than one candidate gene, with some patients possessing five or more such genes (Supplementary Material, Table S4). Thus, where multiple dosage-sensitive genes are affected by CNVs, each may contribute additively, by eliciting specific aspects of the phenotype. Alternatively, multiple gene alleles may act combinatorially through shared pathways to yield emergent consequences. This alternative explanation may account for nine genes whose phenotypes are only revealed in compound knockouts that were identified in this study as candidates for DD or secondary phenotypes. Of these nine, all but one (ST8SIA2) lie within CNVs harboring multiple candidate genes (Table 2 and Supplementary Material, Table S4). This provides evidence for epistasis among multiple CNV alleles controlling DD and associated phenotypes.
Secondary phenotypes
Model phenotypes that we identified for these DD patients appear to readily explain their secondary phenotypes. For example, mouse retina abnormalities were associated with human eye abnormalities, and mouse abnormal conditioning behavior was associated with human behavioral abnormalities. However, the specific association of four mouse model phenotypes (abnormal voluntary movement, abnormal stationary movement, abnormal emotion/affect behavior and abnormal response to novelty) with patients with seizures might appear less obvious. These associations appear not to be chance observations as two of these model phenotypes (abnormal stationary movement and abnormal voluntary movement) are replicated in our previously described LD cohort (+174% enrichment, P = 3 × 10−4; and, +59% enrichment, P = 2 × 10−3, respectively), and are also found to be segregating with Gain CNVs (+517% and +273% enrichments, respectively). One possibility we considered is that copy number changes involving multiple genes jointly perturb a shared neurological pathway so as to produce a phenotype, which is distinct from that resulting due to disruptions of each single gene. Indeed, our observation that the median number of seizure candidate genes per seizure patient is 3.5 is consistent with this scenario of mass action and emergent properties (30,37).
CONCLUSIONS
Drawing together our findings, we can now address the five questions that we outlined in the Introduction. (i) We identified two largely non-overlapping enrichments of genes associated with abnormal brain commissure morphologies and of genes associated with abnormal long-term potentiation within DD-associated CNVs. This indicates that at least two different pathoetiologies underlie the DD clinical phenotype. Furthermore, as the candidate genes from these two enrichments together can explain up to 46% of these patient's disorders, further pathoetiologies are likely to be discovered. (ii) For the primary DD phenotype, CNVs harboring a candidate gene contain on average only one such candidate gene, suggesting that only a single functional CNV element is responsible for the primary phenotype. However, for the three secondary symptoms for which we identify enrichments, CNVs with candidate genes possess, on average, two or more candidate genes which raise the possibility that they act in a combinatorial manner. Although further investigation is required to determine genetic interactions, generally we find no need to invoke non-additive effects to explain pathogenesis. However, as discussed above, patients exhibiting (involuntary) seizures show CNV for, on average, 3.5 genes which in mouse are each associated with an abnormal voluntary movement phenotype. In this case, interactions among these genes might explain the inequality between the human symptom and this mouse model phenotype. (iii) Of 52 candidate genes for the primary DD phenotype, 21 (50%) are also candidate genes for secondary symptoms. Our findings thus suggest that a single pathogenic element will often contribute to both primary and secondary patient traits. (iv) We find both quantitative and qualitative differences between Gain and Loss CNVs within our cohort. For DD-associated enrichments, we find that Gain CNVs overlap significantly more candidate genes than Loss CNVs, while secondary symptoms are associated with Gain or Loss CNVs, but never both. (v) The DD cohort and a previously described LD cohort are both significantly associated with an abnormal tract model phenotype. Associations with other model phenotypes, on the other hand, are specific either to LD or to DD.
This study demonstrates how the application of mouse experimental data en masse provides a formidable functional genomics resource. The phenotypic enrichments identified through this approach are more readily interpretable than terms that might be identified through comparable functional genomics resources, such as Gene Ontology (GO) (38). Moreover, in our hands mouse phenotype data are considerably more informative than GO or gene-expression information. DD CNV genes exhibit no significant enrichments of overarching GO (GOSlim) terms, and they are also not significantly enriched in brain-specific expression, using previously described approaches (12). The mouse knockout resource is important in one other respect, namely that disease-relevant mouse models are often readily available for further investigation, either from repositories such as the Jackson Laboratory, or from the International Knockout Mouse Consortium (39).
Furthermore, this study has demonstrated the utility of collectively analyzing detailed phenotypic information from relatively large, patient cohorts diagnosed with CNV-based DD/LD disorders. Approaches such as ours, that computationally exploit the available functional genomic resources, hold great potential for the identification of candidate genes whose alterations affect multiple systems during early human development. Current improvement in the speed, and reduction in costs, of whole genome and exome sequencing have the potential of cheaply generating large datasets from patient cohorts. We can foresee datasets such as ours being invaluable in prioritizing genes and genomic regions for analysis in patients with DD/LD disorders.
MATERIALS AND METHODS
Patient samples
All patient and normal samples used in this study were collected after obtaining informed consent under protocols approved by the Children's Hospital of Philadelphia (CHOP) Institutional Review Board. Genomic DNA was prepared either from peripheral blood lymphocytes (PBLs) or from subject-derived cell lines using the PuregeneTM DNA isolation kit (Gentra Systems Inc. Minneapolis, MN, USA). In six out of the 87 patients reported here, genomic DNA extracted from lymphoblastoid cell lines was used for array analysis; the remaining 81 genomic DNA samples were extracted directly from PBLs.
CNV detection, validation and data analysis
Microarray experiments were performed using Affymetrix GeneChip 500K or SNP 6.0 arrays (Affymetrix, Santa Clara, CA, USA) or Illumina Infinium™ II HumanHap550 BeadChip (Illumina, San Diego, CA, USA). Genomic DNA from the subject was processed and labeled using reagents and protocols supplied by the manufacturers. Affymetrix arrays were analyzed with the Partek Genomics Suite (Partek Inc, St. Louis, MO, USA) for CNV detection, and Illumina arrays were analyzed using BeadStudio 3.0 software package (Illumina) for CNV detection. Detected CNVs were further analyzed and annotated using CNV Workshop (40). All abnormalities detected by microarray analysis were confirmed and visualized either by metaphase fluorescence in situ hybridization (FISH) or real-time quantitative PCR (q-PCR) as described previously (41,42). For the six patient samples in which genomic DNA used for array analysis was obtained from lymphoblastoid cell lines, CNV validation was performed by metaphase FISH on cell pellets that were prepared from PBLs, thus ruling out cell line artifacts. Parental analysis was performed on both parents for each of the patients reported and was used to confirm de novo status of the observed CNVs. The parental analysis was either performed in our laboratory or the parental data were available from the reports of analysis performed in clinical diagnostic laboratories. Parental samples were analyzed using locus-specific assays, either FISH or q-PCR (as above) to validate CNVs observed in patients. Observed CNVs were compared with the available CNV databases including the Database of Genomic Variants (http://projects.tcag.ca/variation/) and a control CNV database generated from over 2000 controls at CHOP (http://cnv.chop.edu/) in order to eliminate those that appear to be common in the healthy general population.
Furthermore, we also removed from consideration those regions overlapped by, and exhibiting the same direction of copy change as, 26 452 control CNVs that we had employed as a control set in a previous analysis (12). This control CNV set was formed from 25 196 CNVs identified in 240 individuals from Redon et al. (9) combined with 1276 inherited CNVs described in Nguyen et al. (10). Together, these apparently benign CNVs represent 429 Mb of unique sequence (14.0% of the NCBI36 human genome assembly; Table 1 and Supplementary Material, Table S1). Genes within these ‘benign’ CNVs were also analyzed as control enrichments in a manner similar to the genes observed in pathogenic CNVs.
CNV intervals were merged into CNVRs when they overlapped by more than 1 bp. CNV sets were also subdivided according to the direction of copy number change (i.e. Gain or Loss; Table 1). Overlapping CNVs were also merged within each of these subdivisions.
Mouse Genome Informatics phenotypes
Human protein-coding genes were assigned to a CNV if they were completely overlapped by the CNV according to information from Entrez genes (43). Phenotype annotations for disruptions of mouse orthologs of these genes were obtained from the Mouse Genome Informatics (MGI) resource (http://www.informatics.jax.org, version 3.54) (44–46). Specifically, phenotypic associations listed in the MGI file ‘MGI_PhenoGenoMP.rpt’ were mapped on to the human Entrez genes listed in ‘HMD_HumanPhenotype.rpt’. Annotations assigned to genes by the MGI resource represent (i) only the most specific phenotypes that have been reported within a published experiment, and (ii) the over-arching phenotypic category under which those phenotypes fall (Paul Szauter [MGI], personal communication). Thus, the MGI resource might report a highly specific (fine level) term (e.g. abnormal pars anterior morphology) and a general (coarse level) phenotypic category (i.e. nervous system) but not intervening terms (e.g. abnormal tract) that are linked through parent–child relationships within the Mammalian Phenotype Ontology (11). Consequently, and in contrast to our previous analysis (12), we developed the method to allow it to consider not only phenotypes supplied by the MGI resource but also the imputed linking terms between them within the ontology.
Using this approach and taking advantage of simple, unambiguous, 1:1 gene orthology relationships from the MGI resource, 5329 distinct MGI phenotypic terms were mapped to 5011 human genes. Thereafter, we considered only those phenotypic terms present within the well-populated nervous system phenotypes, defined as those terms associated with at least 1% of all genes annotated with any nervous system phenotype. This resulted in the investigation of 146 phenotypic terms that were associated with 1804 mouse–human orthologs. This reduction of phenotypic terms under consideration limits poorly populated and therefore uninformative results, and reduces the number of tests performed thereby improving the method's power. We then tested, using the hypergeometric test, the null hypothesis that a (mouse) phenotype associated with (human) Entrez genes overlapping a set of DD-associated genomic intervals occurs at a frequency that is no different from that obtained by random sampling of all 5011 human genes whose mouse orthologs have a documented phenotypes when disrupted. False discovery observations were controlled by applying an FDR threshold of <5% (47) (see below). Our approach makes the reasonable assumption that MGI mouse phenotypes have been annotated independently of whether their associated human genes lie inside or outside of the DD-associated CNVs. Given that a large number of phenotypic terms were being tested, and that the assumption of independence between terms when applying an FDR correction is unrealistic, application of this significance threshold is likely to be highly conservative.
Linking model phenotypes to patient secondary phenotypes
Many of the patients considered in this study present clinical features in addition to DD. For each additional clinical feature, such as seizures or brain malformations, we were interested in testing for significant enrichments of mouse model phenotypes associated with these patients’ CNV genes. Patients were grouped on the basis of 12 secondary clinical features (Supplementary Material, Table S2).
Phenotypic data were collected from clinical reports based on examination by a team of highly experienced geneticists, dysmorphologists, neurologists and other pediatric specialists at CHOP. Secondary phenotypes such as dysmorphic facial features, cleft lip, cleft palate, sensorineural hearing loss, seizure disorders and short stature were based on these clinical assessments and observations. Behavioral abnormalities recorded in our patient cohort included autistic spectrum disorders, Asperger's syndrome, Attention-deficit/hyperactivity disorder, tantrums, mood disorder, anger, aggression, head banging and biting. Brain malformations recorded in our patient cohort included craniosynostosis, microcephaly, plagiocephaly, brachycephaly, agenesis of corpus callosum, absent right cerebellar hemisphere, abnormal magnetic resonance imaging findings, immature brain formation and delayed myelination. Cardiac defects recorded in our patient cohort included atrial septal defects, ventricular septal defects, truncus arteriosus, tetralogy of fallot, non-compaction cardiomyopathy, coarctation of the aorta, pulmonary stenosis and patent ductus arteriosus. Eye abnormalities included retinoblastoma, hamartoma, strabismus and nystagmus. Limb abnormalities included digital abnormalities and small hands and feet. Urogenital abnormalities included abnormal genital development, XY karyotype with female genitalia, webbed penis, ureteropelvic junction obstruction, urinary reflux and unilateral hydronephrosis.
All CNVs from patients exhibiting a particular clinical feature were then merged into CNVRs as before. CNVRs were also assembled separately from Loss or Gain CNVs into Loss-only and Gain-only CNVRs, respectively. In a similar manner, the CNVs of patients that did not exhibit the particular secondary clinical feature were assembled to form control CNVR sets. Tests were limited to only those categories of secondary model phenotypes that were considered a priori as being relevant to each of these 12 secondary clinical features (Supplementary Material, Table S2). As before, we considered only those phenotypes populated with >1% of all genes annotated within the phenotypic category. Genes overlapped by these CNVRs were then examined for enrichments of these phenotypic terms. An FDR upper threshold of 5% was applied to all P-values, as before, in order to control the rate of false discoveries.
Statistical tests
The significance of enrichments or deficits of genes associated with particular mouse model phenotypes was evaluated using the hypergeometric test that describes the number of successes in a sequence of x draws from a finite population without replacement. More specifically, considering only those 5011 human genes with a mouse ortholog whose disruption had been phenotyped and given the proportion of these that possessed a particular phenotype (for example, see Table 2), we calculated the likelihood of obtaining the observed number of genes with that particular phenotype (for example, see Table 2) simply by chance among those genes overlapped by a given set of CNVs (Table 1). For example, given the total population of 5011 human genes with disrupted and phenotyped mouse orthologs, of which 71 non-exclusively yield a reduced long-term potentiation phenotype (Table 2), the likelihood of a random sample of 808 genes containing 24 genes whose disrupted mouse ortholog yields a reduced long-term potentiation phenotype is 1.8 × 10−4 (Table 2). Where multiple tests were performed, the application of an FDR multiple testing correction was applied to ensure a less than 5% likelihood of any significant term being a false-positive (47).
Calculation of the fold-enrichment within the DD-associated CNVs for the final set of 52 DD-associated candidate genes was performed by random sampling. One thousand gene sets, matched in gene number to that within the DD-associated CNVRs, were obtained by random sampling and the median expected number of genes, 28, annotated with one or more of the significantly enriched terms (Fig. 1) were recorded. Given the 52 candidate genes within the DD-associated CNVRs, we thus estimate an ∼1.9-fold enrichment over the number expected by chance.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institutes of Health (GM081519 to T.H.S., 5-T32-GM-008638-11 to C.H.-E.). This work was supported by Medical Research Council, UK funding (C.P.P. and C.W.) and by the European Union's Seventh Framework Program under grant agreement no. (241995), project GENCODYS (C.P.P. and C.W.).
ACKNOWLEDGEMENTS
We are very grateful to patients and their families for participating in this study. We would also like to thank all the clinicians and genetic counselors who enrolled their patients into our study, especially Elaine Zackai, Sulagna Saitta, Carsten Bonneman, Donna McDonald-McGinn, Livija Medne and their colleagues at the Children's Hospital of Philadelphia.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. doi:10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 2.Pober B.R. Williams-Beuren syndrome. N. Engl. J. Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. doi:10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 3.Gothelf D., Frisch A., Michaelovsky E., Weizman A., Shprintzen R.J. Velo-Cardio-Facial syndrome. J. Ment. Health. Res. Intellect. Disabil. 2009;2:149–167. doi: 10.1080/19315860902756136. doi:10.1080/19315860902756136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elsea S.H., Girirajan S. Smith-Magenis syndrome. Eur. J. Hum. Genet. 2008;16:412–421. doi: 10.1038/sj.ejhg.5202009. doi:10.1038/sj.ejhg.5202009. [DOI] [PubMed] [Google Scholar]
- 5.Slavotinek A.M. Novel microdeletion syndromes detected by chromosome microarrays. Hum. Genet. 2008;124:1–17. doi: 10.1007/s00439-008-0513-9. doi:10.1007/s00439-008-0513-9. [DOI] [PubMed] [Google Scholar]
- 6.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh036. doi:10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. doi:10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P., et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. doi:10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen D.Q., Webber C., Hehir-Kwa J.Y., Pfundt R., Veltman J.A., Ponting C.P. Reduced purifying, not positive, selection explains genomic bias amongst copy number variation. Genome Research. 2008;18:1711–1723. doi: 10.1101/gr.077289.108. doi:10.1101/gr.077289.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith C.L., Goldsmith C.A., Eppig J.T. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. doi:10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webber C., Hehir-Kwa J.Y., Nguyen D.Q., de Vries B.B., Veltman J.A., Ponting C.P. Forging links between human mental retardation-associated CNVs and mouse gene knockout models. PLoS Genet. 2009;5:e1000531. doi: 10.1371/journal.pgen.1000531. doi:10.1371/journal.pgen.1000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akil H., Brenner S., Kandel E., Kendler K.S., King M.C., Scolnick E., Watson J.D., Zoghbi H.Y. Medicine. The future of psychiatric research: genomes and neural circuits. Science. 2010;327:1580–1581. doi: 10.1126/science.1188654. doi:10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W., Quintero-Rivera F., Fan Y., Alkuraya F.S., Donovan D.J., Xi Q., Turbe-Doan A., Li Q.G., Campbell C.G., Shanske A.L., et al. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet. 2007;3:e80. doi: 10.1371/journal.pgen.0030080. doi:10.1371/journal.pgen.0030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi: 10.1086/320609. doi:10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motazacker M.M., Rost B.R., Hucho T., Garshasbi M., Kahrizi K., Ullmann R., Abedini S.S., Nieh S.E., Amini S.H., Goswami C., et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 2007;81:792–798. doi: 10.1086/521275. doi:10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boycott K.M., Flavelle S., Bureau A., Glass H.C., Fujiwara T.M., Wirrell E., Davey K., Chudley A.E., Scott J.N., McLeod D.R., et al. Homozygous deletion of the very low density lipoprotein receptor gene causes autosomal recessive cerebellar hypoplasia with cerebral gyral simplification. Am. J. Hum. Genet. 2005;77:477–483. doi: 10.1086/444400. doi:10.1086/444400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown S.A., Warburton D., Brown L.Y., Yu C.Y., Roeder E.R., Stengel-Rutkowski S., Hennekam R.C., Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 1998;20:180–183. doi: 10.1038/2484. doi:10.1038/2484. [DOI] [PubMed] [Google Scholar]
- 19.Dalski A., Atici J., Kreuz F.R., Hellenbroich Y., Schwinger E., Zuhlke C. Mutation analysis in the fibroblast growth factor 14 gene: frameshift mutation and polymorphisms in patients with inherited ataxias. Eur. J. Hum. Genet. 2005;13:118–120. doi: 10.1038/sj.ejhg.5201286. doi:10.1038/sj.ejhg.5201286. [DOI] [PubMed] [Google Scholar]
- 20.Paul L.K., Brown W.S., Adolphs R., Tyszka J.M., Richards L.J., Mukherjee P., Sherr E.H. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat. Rev. Neurosci. 2007;8:287–299. doi: 10.1038/nrn2107. doi:10.1038/nrn2107. [DOI] [PubMed] [Google Scholar]
- 21.Bedeschi M.F., Bonaglia M.C., Grasso R., Pellegri A., Garghentino R.R., Battaglia M.A., Panarisi A.M., Di Rocco M., Balottin U., Bresolin N., et al. Agenesis of the corpus callosum: clinical and genetic study in 63 young patients. Pediatr. Neurol. 2006;34:186–193. doi: 10.1016/j.pediatrneurol.2005.08.008. doi:10.1016/j.pediatrneurol.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Sherr E.H., Owen R., Albertson D.G., Pinkel D., Cotter P.D., Slavotinek A.M., Hetts S.W., Jeremy R.J., Schilmoeller G., Schilmoeller K., et al. Genomic microarray analysis identifies candidate loci in patients with corpus callosum anomalies. Neurology. 2005;65:1496–1498. doi: 10.1212/01.wnl.0000183066.09239.b6. doi:10.1212/01.wnl.0000183066.09239.b6. [DOI] [PubMed] [Google Scholar]
- 23.Sylvester P.E. The anterior commissure in Down's syndrome. J. Ment. Defic. Res. 1986;30(Pt 1):19–26. doi: 10.1111/j.1365-2788.1986.tb01294.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuker W., Mayrhofer H., Mader I., Nagele T., Krageloh-Mann I. Malformations of the midline commissures: MRI findings in different forms of callosal dysgenesis. Eur. Radiol. 2003;13:598–604. doi: 10.1007/s00330-002-1541-9. [DOI] [PubMed] [Google Scholar]
- 25.Schell-Apacik C.C., Wagner K., Bihler M., Ertl-Wagner B., Heinrich U., Klopocki E., Kalscheuer V.M., Muenke M., von Voss H. Agenesis and dysgenesis of the corpus callosum: clinical, genetic and neuroimaging findings in a series of 41 patients. Am. J. Med. Genet. A. 2008;146A:2501–2511. doi: 10.1002/ajmg.a.32476. doi:10.1002/ajmg.a.32476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mowat D.R., Wilson M.J., Goossens M. Mowat-Wilson syndrome. J. Med. Genet. 2003;40:305–310. doi: 10.1136/jmg.40.5.305. doi:10.1136/jmg.40.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer G.B., Bodensteiner J.B., Buehler B.A., Lin A., Cole T.R. The neuroimaging findings in Sotos syndrome. Am. J. Med. Genet. 1997;68:462–465. doi: 10.1002/(sici)1096-8628(19970211)68:4<462::aid-ajmg18>3.0.co;2-q. doi:10.1002/(SICI)1096-8628(19970211)68:4<462::AID-AJMG18>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 28.Battaglia F., Quartarone A., Rizzo V., Ghilardi M.F., Di Rocco A., Tortorella G., Girlanda P. Early impairment of synaptic plasticity in patients with Down's syndrome. Neurobiol. Aging. 2008;29:1272–1275. doi: 10.1016/j.neurobiolaging.2007.02.025. doi:10.1016/j.neurobiolaging.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Vaillend C., Poirier R., Laroche S. Genes, plasticity and mental retardation. Behav. Brain Res. 2008;192:88–105. doi: 10.1016/j.bbr.2008.01.009. doi:10.1016/j.bbr.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Galdzicki Z., Siarey R., Pearce R., Stoll J., Rapoport S.I. On the cause of mental retardation in Down syndrome: extrapolation from full and segmental trisomy 16 mouse models. Brain Res. Brain. Res. Rev. 2001;35:115–145. doi: 10.1016/s0926-6410(00)00074-4. doi:10.1016/S0926-6410(00)00074-4. [DOI] [PubMed] [Google Scholar]
- 31.Mason S., Piper M., Gronostajski R.M., Richards L.J. Nuclear factor one transcription factors in CNS development. Mol. Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. doi:10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 32.Wang W., Mullikin-Kilpatrick D., Crandall J.E., Gronostajski R.M., Litwack E.D., Kilpatrick D.L. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J. Neurosci. 2007;27:6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. doi:10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barry G., Piper M., Lindwall C., Moldrich R., Mason S., Little E., Sarkar A., Tole S., Gronostajski R.M., Richards L.J. Specific glial populations regulate hippocampal morphogenesis. J. Neurosci. 2008;28:12328–12340. doi: 10.1523/JNEUROSCI.4000-08.2008. doi:10.1523/JNEUROSCI.4000-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein R. Bidirectional modulation of synaptic functions by Eph/ephrin signaling. Nat. Neurosci. 2009;12:15–20. doi: 10.1038/nn.2231. doi:10.1038/nn.2231. [DOI] [PubMed] [Google Scholar]
- 35.Samuels I.S., Saitta S.C., Landreth G.E. MAP'ing CNS development and cognition: an ERKsome process. Neuron. 2009;61:160–167. doi: 10.1016/j.neuron.2009.01.001. doi:10.1016/j.neuron.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Swieten J.C., Brusse E., de Graaf B.M., Krieger E., van de Graaf R., de Koning I., Maat-Kievit A., Leegwater P., Dooijes D., Oostra B.A., et al. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia [corrected] Am. J. Hum. Genet. 2003;72:191–199. doi: 10.1086/345488. doi:10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki G., Harper K.M., Hiramoto T., Funke B., Lee M., Kang G., Buell M., Geyer M.A., Kucherlapati R., Morrow B., et al. Over-expression of a human chromosome 22q11.2 segment including TXNRD2, COMT and ARVCF developmentally affects incentive learning and working memory in mice. Hum. Mol. Genet. 2009;18:3914–3925. doi: 10.1093/hmg/ddp334. doi:10.1093/hmg/ddp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium . Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. doi:10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins F.S., Rossant J., Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. doi:10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Gai X., Perin J.C., Murphy K., O'Hara R., D'Arcy M., Wenocur A., Xie H.M., Rappaport E.F., Shaikh T.H., White P.S. CNV Workshop: an integrated platform for high-throughput copy number variation discovery and clinical diagnostics. BMC Bioinform. 2010;11:74. doi: 10.1186/1471-2105-11-74. doi:10.1186/1471-2105-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ming J.E., Geiger E., James A.C., Ciprero K.L., Nimmakayalu M., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., et al. Rapid detection of submicroscopic chromosomal rearrangements in children with multiple congenital anomalies using high density oligonucleotide arrays. Hum. Mutat. 2006;27:467–473. doi: 10.1002/humu.20322. doi:10.1002/humu.20322. [DOI] [PubMed] [Google Scholar]
- 42.Shaikh T.H., Gai X., Perin J.C., Glessner J.T., Xie H., Murphy K., O'Hara R., Casalunovo T., Conlin L.K., D'Arcy M., et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. doi:10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. doi:10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Blake J.A. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–728. doi: 10.1093/nar/gkm961. doi:10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eppig J.T., Blake J.A., Bult C.J., Richardson J.E., Kadin J.A., Ringwald M. Mouse genome informatics (MGI) resources for pathology and toxicology. Toxicol. Pathol. 2007;35:456–457. doi: 10.1080/01926230701310536. doi:10.1080/01926230701310536. [DOI] [PubMed] [Google Scholar]
- 46.Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., et al. The Mouse Genome Database (MGD): from genes to mice–a community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–475. doi: 10.1093/nar/gki113. doi:10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., Ser. B (Methodol.) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.