Abstract
Background
The genetic analysis of human primary immunodeficiencies has defined the contribution of specific cell populations and molecular pathways in host defense against infections. Disseminated infection caused by BCG vaccines is an early manifestation of primary immunodeficiencies, such as severe combined immunodeficiency. In many affected individuals, the etiology of disseminated BCG disease is unexplained.
Methods
We investigated an infant presenting with features of severe immunodeficiency, including early-onset disseminated BCG disease, requiring hematopoietic stem cell transplantation. We also studied two otherwise healthy adults with a history of disseminated but curable BCG disease in childhood. We characterized the monocyte and dendritic cells compartments in these three persons and sequenced candidate genes, mutation of which could plausibly confer susceptibility to BCG disease.
Results
We detected two distinct disease-causing mutations affecting the transcriptional regulator IRF8. Both K108A and T80A mutations impair IRF8 transcriptional activity by disrupting IRF8 interaction with DNA. Mutation K108E was associated with an autosomal recessive severe immunodeficiency with a complete lack of circulating monocytes and dendritic cells. Mutation T80A was associated with an autosomal dominant milder immunodeficiency and a selective depletion of CD11c+ CD1c+ circulating dendritic cells.
Conclusions
These findings define a new class of human primary immunodeficiency, affecting the differentiation of mononuclear phagocytes. They also demonstrate that human IRF8 is critically required for the development of monocytes and dendritic cells and for anti-mycobacterial immunity.
INTRODUCTION
The discovery of human primary immunodeficiencies (PIDs) affecting the development of granulocytes, B cells, and T cells, has been instrumental in defining the contribution of these cell types to protective immunity[1,2]. Monocytes, macrophages, and dendritic cells — all mononuclear phagocytes — have essential functions in both innate and acquired immunity. They initially recognize and engulf invading microbes, produce pro-inflammatory cytokines (such as IL-12), and process antigens for presentation to naïve T lymphocytes, which consequently secrete various lymphokines (such as IFN-γ)[3,4]. Upon activation by cytokines secreted by T lymphocytes, mononuclear phagocytes destroy ingested micro-organisms. There are no known genetic causes of PID affecting the development of the mononuclear phagocyte.
BCG vaccines and environmental mycobacteria are efficiently destroyed by T cell-activated macrophages[5]. However, disseminated mycobacterial disease following BCG vaccination is a sign of immunodeficiency[6]. It occurs in children with severe combined immunodeficiency (absence of T lymphocytes) or with chronic granulomatous disease. While such children are vulnerable to multiple infections, individuals with Mendelian Susceptibility to Mycobacterial Disease (MSMD)[7] display a narrow vulnerability to poorly virulent mycobacteria, including BCG. Some MSMD individuals harbor genetic defects in the IL-12/IFNγ circuit, with mutations in genes encoding IL-12p40 (encoded by IL12B), and its receptor, both subunits of the receptor for IFNγ, the IFN-γR signaling partner STAT1, the NF-κB regulator Iκκ̃γ (NEMO), and the effector gp91phox[7].
We hypothesized that certain individuals may be vulnerable to BCG because of mutations impairing the development of mononuclear phagocytes. Among the genes encoding transcription factors that are essential to the development of mononuclear phagocytes in mice, IRF8, the interferon regulatory factor IRF8 stood out as a strong candidate[8]. It is expressed at very high levels in mononuclear phagocytes[9], and regulates granulocyte/macrophage differentiation[10,11] and DC development[12–15]. Acting in heterodimeric complexes with other transcription factors, IRF8 also controls the transcriptional response of mature myeloid cells to interferons (IFNs) and Toll-like receptor (TLR) agonists, a response in which IRF8 binds and trans-activates the promotors of IL12B and NOS2.[8] (NOS2 encodes inducible nitric oxide synthase iNOS.) Mice bearing the R294C hypomorphic mutation (a mutation that partially compromises protein function) in Irf8 display a specific DC phenotype with loss of CD8α+ lymphoid DCs and CD103+ tissue myeloid DCs, while Irf8 knockout mice (which are devoid of Irf8) additionally lack plasmacytoid DCs[13–15]. Mice mutated in irf8 are susceptible to infection with intra-macrophagic pathogens, and are hyper-susceptible to M. bovis BCG and M. tuberculosis infections[16,17].
METHODS
Study Subjects
A 10-week old infant presenting with disseminated BCG infection (post-vaccination), oral candidiasis, cachexia and suspected signs of immunodeficiency was admitted to hospital and was investigated (Case Description in Supplementary Information). She underwent multiple rounds of aggressive antibiotic treatments which were only partially effective in improving her health. She was successfully treated by transplantation wtih cord-blood stem cells. She was the second-born child to healthy unrelated Irish parents (kindred A); the elder sibling was well, and all had received BCG vaccination.
Two additional individuals diagnosed with MSMD were also enrolled in the study. Affected individual 1 (kindred B) was born in 1970 to non-consanguineous parents of Italian descent living in Brazil. He was vaccinated with BCG at birth and developed recurrent episodes of lymphadenopathies at age 15 months, 20 years and 30 years; on two such episodes, lymph node biopsies revealed presence of acid-fast bacilli and all such episodes were successfully treated with anti-mycobacterial drug regimens. His mother, father and sister were vaccinated with BCG and did not present with clinical infectious disease suggesting immunodeficiency.
Affected individual 2 (kindred C) was born in 1996 to a non consanguineous family of Italian descent living in Chile, and received BCG vaccination at birth. At one year of age, she developed lymphadenopathy with chronic granulomatous tuberculoid lesions. At two years of age, she presented with multiple lymphadenopathies and fever, requiring hospital admission. Histological analysis of lymph-node biopsy and bacterial culture identified pyrazinamide resistant M. bovis, and the child was successfully treated with adjusted antibiotic treatment administered for 12 months; no subsequent clinical episodes were reported. We obtained written informed consent from participants or their parents, and Institutional Review Boards approved the studies.
Genetic and Transcriptional Analyses
We sequenced the exons and selected non-coding sequences of IRF8 from participants’ genomic DNA following standard PCR amplification with sequence-specific primers (Table S3). We assayed the transcriptional activity of mutant and non-mutant IRF8 through activation of the IL12B and NOS2 promoters using reporter constructs, transiently transfected into RAW 264.7 macrophages, as previously described[16].
Biochemical Assays, Molecular Characterization and Statistical Analysis
See the Supplementary Appendix for details.
RESULTS
Autosomal Recessive IRF8 Deficiency
We evaluated a 10-week-old infant presenting with disseminated BCG disease (post-vaccination), cachexia and oral candidiasis (see Supplementary Appendix). The blood count revealed a strikingly abnormal myeloid compartment with absence of monocytes and basophils and a very high neutrophil count (Fig. 1; Table S1). Examination of peripheral blood using flow cytometry confirmed severe depletion of the CD3/19/56− (lin−) HLA-DR+ cellular compartment, with total absence of both CD14+ and CD16+ monocytes (Fig. 1A–C). Furthermore, we could not detect DCs in the blood, whether CD11c+ myeloid DCs (CD1c+ or CD141+) or CD123+ plasmacytoid DCs (Fig. 1A–D). The only lin−DR+ cells proved to be circulating CD34+ progenitor cells, which were elevated (Fig. S1, Panel A) and correlated with elevated serum levels of Fms-like tyrosine kinase-3 ligand (Flt3L)(Fig. S1, Panel B). In contrast, B and NK cells were present in normal numbers (Table S2 and Fig. S2), thus excluding the recently described syndrome of autosomal dominant and sporadic monocytopenia[18,19].
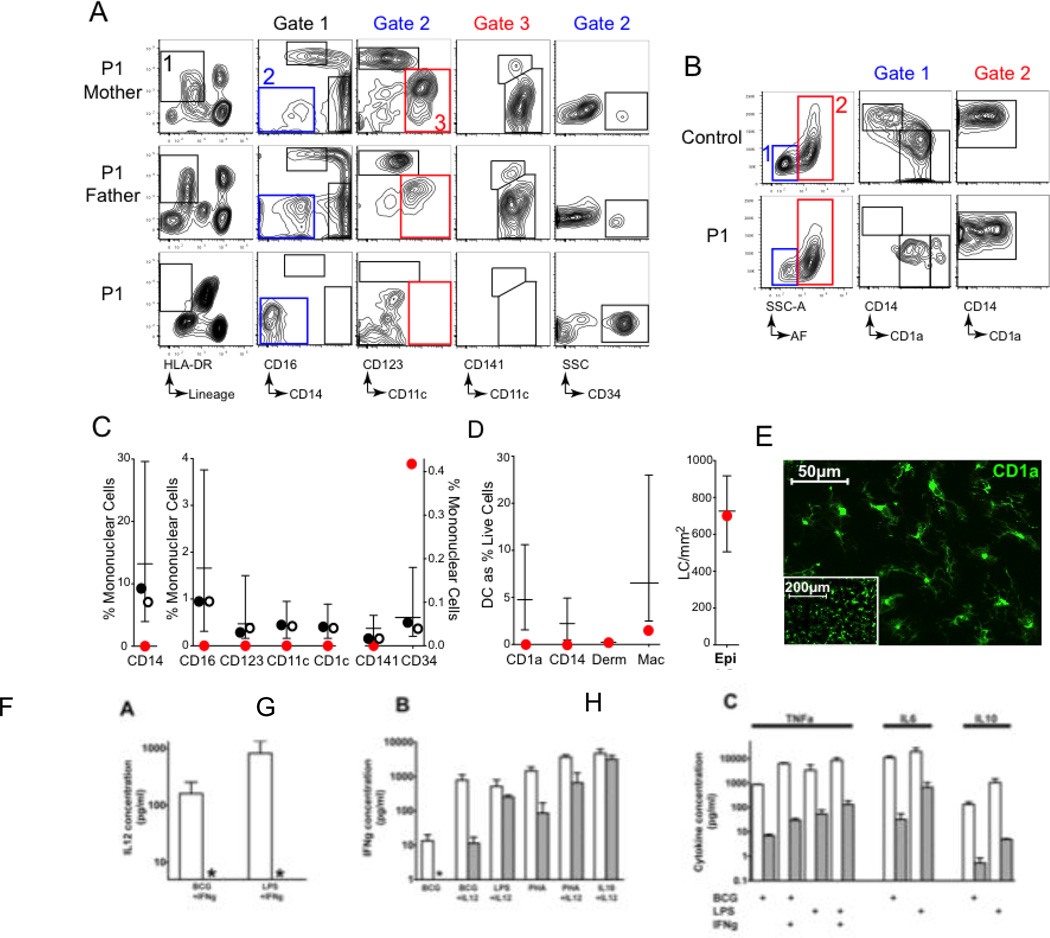
Figure 1. Severe depletion of the antigen presenting cell (APC) compartment in autosomal recessive IRF8 deficiency.
(A)Flow cytometric evaluation of peripheral blood of affected individual P1 and parents. (B) Flow cytometric evaluation of CD45+HLA-DR+ mononuclear cells from collagenase digested dermis of P1 and control. (C) Quantification of peripheral blood APC subsets as % of mononuclear cells in P1 (red circle), mother (filled circle) and father (open circle) compared with normal adult range (box and whiskers: mean and range; n=28). (D) Quantification of dermal APC subsets as % of live cells, compared with normal adult range (box and whiskers: mean and range; n=22) and Langerhans cells per mm2 compared with normal range (box and whiskers: mean and range; n=12). (E) Affected P1 Langerhans Cells (LC) in an epidermal sheet (×400 magnification; inset × 100), revealed by anti-CD1a immunofluorescence.(F–H) Whole blood cytokine responses measured in vitro. (F) IL-12 production in response to BCG or LPS plus exogenous IFNγ. (G) IFNγ production elicited by indicated stimuli. (H) Production of TNFα, IL-6 and IL-10 in response to indicated combinations of BCG, LPS and exogenous IFNγ. Control, empty bars; affected individual, filled bars; *, undetectable.
Through assays using whole blood, we observed that IL12 production in response to BCG, phytohaemagglutinin (PHA) and lipopolysaccharide was completely absent and IFN-γ production was poor, with similarly poor production of TNF-α, IL-10 and IL-6 (Fig.1F–H). Preincubation of the infant’s cells with IL-12 could partially restore IFN-γ production in response to the same stimuli (Fig. 1G). Biopsies of the infant's bone marrow and axillary lymph node (Fig. S3) showed striking myeloid hyperplasia, and the presence of acid-fast bacilli within granulomata in (a) lymph node. Despite the subject’s profound peripheral monocytopenia, the diseased node was positive for histiocytic markers CD68 and CD163 and bone trabeculae showed evidence of normal osteoclast activity (Fig. S4). In dermal tissues, the density of CD1a+ and CD14+ DCs was remarkably low (Fig. 1C–D). In contrast, we observed epidermal Langerhans cells (LCs) to be at normal density (Fig. 1E), and detected the occasional Langerin+/CD1a+ DC in lymph node (Fig. S4). We conclude that the subject had a profound deficit of tissue DCs, blood DCs and monocytes, a variable deficit of tissue macrophages, and normal levels of LCs.
This combination of immunodeficiency [10,11], DC deficiency [12–15] and myeloproliferation [20] is strikingly similar to the phenotype of mice harboring loss-of-function mutations in Irf8. On sequencing IRF8 in the subject, we observed a homozygous missense variant predicted to effect a lysine-to-glutamic acid substitution at position 108 (K108E) (Fig. 2A). Both parents were heterozygous for K108E and an unaffected sibling lacked the variant allele (Fig. 2C). We sequenced IRF8 in 454 unrelated persons with clinical susceptibility to mycobacterial infection and did not detect the variant or any other homozygous variants.
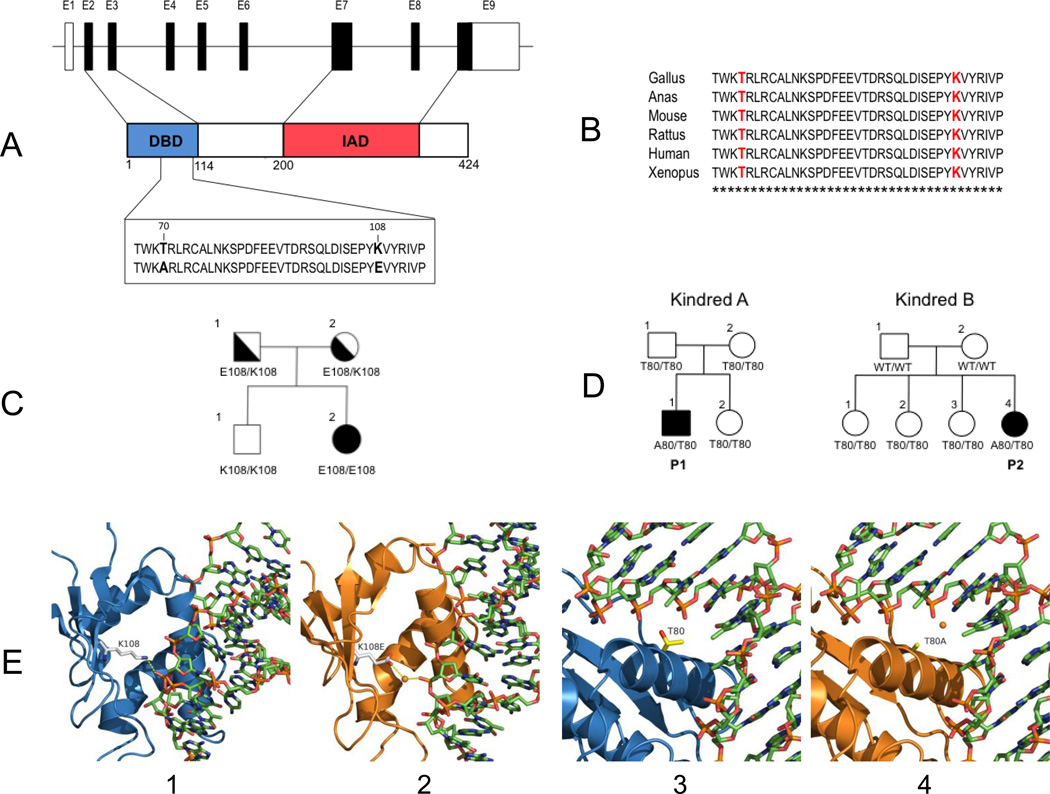
Figure 2. Genetic analysis of autosomal recessive (AR) and autosomal dominant (AD) IRF8 deficiency.
(A) Schematic representation of the IRF8 gene, including the 9 non-coding (empty) and coding (filled) exons, with the major structural features of the protein shown (DBD, DNA Binding Domain; IAD, IRF Association Domain). The position of the T80A and K108E mutations (bold) in the DBD is shown. (B) Multiple sequence alignment of IRF8 from different species (T80 and K108 are shown in red; stars identify invariant residues). (C) Segregation of K108E in the proband family. (D) T80A is a de novo mutation that has arisen independently in two unrelated MSMD families from Chile and Brazil. (E) Molecular modeling of wild type K108 (panel 1), and T80 (panel 3) and mutant variants E108 (panel 2) and A80 (panel 4) on the three-dimensional structure of DNA-bound IRF8. The hydrogen bond formed between the side chain amino group of K108 and the sugar backbone of DNA is interrupted by E108. The replacement of Thr80 with an Ala residue in the key DNA-binding helix (inserted into the major groove) alters the hydrophobic interface between the protein and the DNA.
Amino-acid position 108 is within the DNA-binding domain of IRF8 (Fig. 2A); the lysine residue is invariant in IRF8 orthologs (Fig. 2B) and is highly conserved among human IRF family members (data not shown). We expressed the mutant IRF8 in cultured mouse macrophages. Immunoblotting studies showed similar expression levels of normal (K108) and mutant (K108E) variants, and a comparatively slower electrophoretic mobility of the mutant variant (Fig. 3E), suggesting that the mutation affects overall protein structure/folding. We tested the ability of normal and mutant isoforms to activate transcription of IRF8 targets — the promoters of IL12B (Fig. 3A) and NOS2 (Fig. 3B) — in mouse macrophages. Although the combination of normal IRF8 and its co-activator IRF1 induced a dose-dependent stimulation of IL12B and NOS2 promoters, the K108E variant was almost inactive (Fig. 3A, 3B), suggesting that K108E abrogates the IRF1-dependent transcriptional activity of IRF8. Moreover, the mutant IRF8 variant bound the IL12B promoter far more weakly than did the normal variant (Fig. 3D).
Figure 3. Functional characterization of the mutant IRF8 variants.
(A, B) RAW macrophages were transiently transfected with various amounts (indicated) of the IRF8 variants, with and without IRF1, and IRF8-dependent transactivation potential was monitored with IL12p40 (A) or iNOS (B) promoter constructs linked to a luciferase reporter gene. (C) The ability of T80A and R294C to suppress transactivation of the iNOS promotor by WT IRF8 was evaluated by co-transfecting RAW macrophages with various amounts of the T80A and R294C variants and fixed amounts of WT IRF8. (D) The relative binding affinity of IRF8 variants for the IL12p40 promoter was estimated by chromatin imunoprecipitation in transfected RAW macrophages. (E) Western blot analysis of cell extracts from RAW macrophages expressing wild-type, or the T80A or the K108E mutant IRF8 variants, with detection based on an HA epitope tag inserted in-frame in all constructs.
Molecular modeling using the structure of DNA-bound IRF2 [21] places K108 in a short β strand that runs parallel to the major DNA-binding α helix, suggesting that it makes a critical hydrogen bond with the DNA sugar backbone (3.3A) that facilitates IRF8 docking onto DNA (Fig. 2E-1). Replacement of a positively-charged amino acid (lysine) by one that is negatively charged (glutamic acid) at position 108 would likely prevent the formation of a hydrogen bond (Fig. 2E-2), and cause local repulsion of the supporting IRF8 β strand away from DNA, allowing a water molecule to fill the space. This model predicts loss of DNA binding and thus loss of transactivation.
These findings show that the subject carries a loss-of-function mutation in IRF8. The heterozygous parents were healthy despite having received BCG as neonates, and had normal numbers of peripheral blood monocytes and DCs (Fig. 1), consistent with the autosomal recessive (AR) inheritance of the IRF8K108E allele.
Autosomal Dominant IRF8 Deficiency
In parallel with our analysis of the infant with recessive disease, we sequenced IRF8 in 454 persons with MSMD of unknown genetic etiology (we ruled out the possibility that these persons carried mutations in several of the genes in which alterations are known to cause MSMD; see Supplementary Appendix). Two unrelated persons from Chile and Brazil and suffering from recurrent episodes of disseminated BCG disease, were each found to carry the same de novo heterozygous mutation, predicted to effect a threonine-to-alanine substitution at position 80 (T80A) of IRF8 (Fig. 2A,D). We genetically confirmed reported biological paternity. That each mutation was de novo suggests that the same T80A allele arose independently on two occasions. We did not detect the T80A variant in 1064 healthy controls [41] of diverse relevant ancestry (See Supplementary Appendix). Amino-acid position 80 of IRF8 is within the DNA-binding domain (Fig. 2A); the threonine residue is strictly conserved across IRF8 orthologs (Fig. 2B) and human paralogous genes (data not shown).
The T80A mutation had no effect on level and stability of IRF8 in immortalized B cell lines derived from the subjects (Fig. 3E), or following expression in transfected macrophages (data not shown). When tested for its ability to transactivate the promoters of IL12B (Fig. 3A) and NOS2 (Fig. 3B) in mouse macrophages, the T80A mutant showed relatively low activity (Fig. 3A, 3B), similar to that of the hypomorphic R294C Irf8 variant of Irf8-deficient BXH2 mutant mice (Fig. 3A, 3B). The T80A mutant showed weak recruitment to the IL12B promoter (20% of the amount of the non-mutant IRF8 protein), suggesting that the mutation interferes with DNA binding (Fig. 3D).
Homology modeling shows that T80 maps to the DNA-binding α helix of IRF8 that fits into the major groove, with its side-chain directed towards the DNA bases (Fig. 2E-3). Molecular dynamics simulations indicate that T80A alters the hydrophobic interface between the protein and DNA, and possibly modulates DNA-binding specificity of IRF8 (Fig. 2E-4). Alanine has a smaller side chain than does threonine, and so may allow entry of a water molecule at the DNA:IRF8 interface. Although the functional data suggest that T80A is severely hypomorphic, we investigated whether the T80A variant is negatively dominant (that is, it interferes with the function of the non-mutant IRF8 protein). We co-expressed the mutant and non-mutant alleles in macrophages (Fig. 3C) and observed decreasing activation of the IL12B (not shown) and NOS2 (Fig. 3C) promoters with increasing levels of mutant (T80A) IRF8 against fixed levels of non-variant IRF8. The effect was T80A-specific; we did not observe it to be associated with K108E (data not shown) or R294C mutant proteins. We conclude that the T80A mutation has a dual effect on IRF8 function and causes autosomal-dominant MSMD.
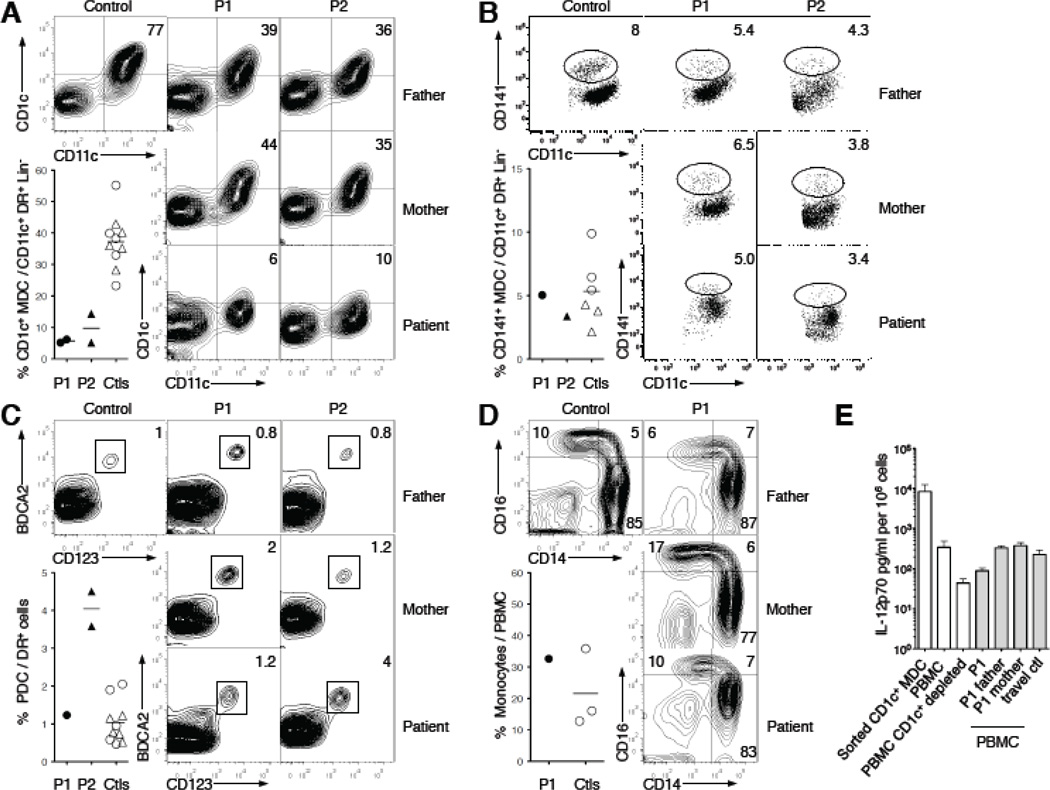
We observed subtle deficits in the peripheral blood mononuclear cells of the two T80A affected adults. There was no deficiency of circulating lymphocytes and granulocytes (data not shown), monocyte subsets, or BDCA2+ CD123+ plasmacytoid DC (Fig. 4D). However, within the CD11c+ myeloid DC subset, which is normally divided into minor CD141+ and major CD1c+ subsets, there was marked loss of CD1c+DCs (Fig. 4A), while the CD141+ subset and total number of CD11c+DCs remained intact (Fig. 4B). CD1c+ DCs from unaffected persons can produce large amounts of IL-12 when stimulated with the TLR7/8 ligand R848 (compared to PBMC; student t test, p<0.02) whereas their depletion from PMBCs was associated with comparatively low levels of IL-12 production by the remaining PBMCs (p<0.02) (Fig. 4E). PBMCs harboring IRF8T80A produced one-third the amount of IL-12 produced by control cells in response to R848 (p<0.004)(Fig. 4E). We suggest that depletion of IL-12-producing CD1c+ DCs contributes to susceptibility to mycobacterial disease in these individuals. In vitro assays using whole blood from the affected persons showed no detectable impact on production of IFN-γ (Fig. S6), or in the subjects’ PBMCs in response to stimulation by PPD (purified protein derivative), BCG or PHA (Fig. S7), or by PHA-driven T-cell blasts stimulated with IL-12 (data not shown). Production of IL-12 in a whole blood assay in response to BCG (Fig. S6), by monocyte-derived DCs stimulated with CD40L (Fig. S8A), and by EBV-B cells stimulated with phorbol dibutyrate (Fig. S8B) was also normal, indicating that T80A does not cause a generalized defect in IL-12 production.
Figure 4. Selective depletion of dendritic cells in autosomal dominant IRF8 deficiency.
(A) Blood CD1c+ dendritic cells are gated on HLA-DR+ Lin− (CD14− CD16− CD19−) cells, and expression of CD11c+ and CD1c+ is shown. Percentage of DR+ CD11c+ Lin− cells that are positive for CD1c+ is shown for the T80A individuals (P1 ●, P2 ▲), their relatives and travel controls (○,△) (mean—). Numbers on contour plots represent the mean of two independent samples. (B) Blood CD141+ dendritic cells are gated on HLA-DR+ Lin− (CD14− CD16− CD19−) cells, and expression of CD11c and CD141 is shown. Percentage of DR+ CD11c+ Lin− cells that are CD141bright is shown. (C) Plasmacytoid dendritic cells are gated on HLA-DR+ Lin− (CD14− CD16−) cells, and expression of CD123 and BDCA2 is shown. Percentage of DR+ cells that are positive for CD123 and BDCA2 is shown. (D) Monoyctes are gated on HLA-DR+ Lin− (CD2− CD15− CD19− Nkp46−) cells, and expression of CD14 and CD16 is shown. Numbers on contour plots represent the percentage of each subset (CD14+, CD14dim and CD14+CD16+) among total monocytes. Percentage of peripheral blood mononuclear cells (PBMC) that are positive for CD14 or CD16 is shown for P1, his relatives and travel control. (E) Production of IL-12p70 (mean ± SD) in response to R848 by FACS-sorted blood CD1c+ (sorted CD1c+ MDC, n=4), PBMCs depleted of CD1c+ DCs (PBMC CD1c+depleted, n=3)) or mock-depleted (PBMC, n=3) from healthy controls, and PBMCs from P1, his relatives and a control collected at the same time (grey bars).
Because the two healthy heterozygous parents of the infant with autosomal-recessive IRF8 deficiency had normal levels of functional DCs, the marked deficit of CD1c+CD11c+ DCs was probably caused by the dominant-negative effect of the T80A mutant protein.
DISCUSSION
We have described three persons, one with autosomal recessive (AR), and two with autosomal dominant (AD) primary IRF8 deficiency, characterized by a complete or selective loss of mononuclear phagocyte subsets, respectively. The IRF8 mutation underlying the AR disorder (K108E) effects an impairment of DNA binding and trans-activation potential of IRF8. This defect produced a life-threatening pediatric syndrome, characterized by absence of blood monocytes and DCs, myeloproliferation of granulocyte precursors, and severe opportunistic infections, together requiring stem cell transplantation in infancy and suggesting that that DC/monocyte deficiency of any cause will produce susceptibility to various infectious pathogens. On the other hand, AD IRF8 deficiency is associated, in the two persons we investigated, with a heterozygous mutation resulting in a dominant-negative IRF8 allele (T80A) that suppresses the trans-activation potential of non-mutant IRF8 in vitro. The associated syndrome was less severe than that of the child with AR disease, and was characterized by an abnormal peripheral blood myeloid DC phenotype with a marked loss of CD11c+CD1c+ DCs. AD IRF8 deficiency causes selective susceptibility to mycobacterial infections, and represents a novel, albeit rare, etiology of MSMD. Together, these results establish a critical role for IRF8 in the ontogeny of the human mononuclear phagocyte lineage and of circulating monocytes and DCs in particular.
Cultivation of circulating stem cells from the child with autosomal-recessive IRF8 deficiency with growth factors that support granulocyte and monocyte/macrophage colony formation showed that myeloid colonies formed were almost exclusively granulocytic (>98%), thus establishing that IRF8 is critical in the differentiation of myeloid progenitors into monocytes. Irf8-deficiency in mice is also associated with myeloproliferation of granulocyte precursors[10,11,20] and very low levels of circulating monocytes (F. Ginhoux and M. Merad, personal communication). That tissue macrophages and Langerhans cells are well-represented in autosomal-recessive IRF8 deficiency suggests heterogeneity within the mononuclear phagocyte compartment with respect to IRF8-independence and/or potential for local self-renewal [22,23]. Further work will be required to establish the relative capacity of IRF8-deficient human CD34+ progenitors to produce fully functional macrophages and DCs in vitro[24]. Studies in IRF8-deficient mice also show normal numbers of F4/80+ tissue macrophages [P. Gros, unpublished], although these cells are abnormally susceptible to infection with intracellular pathogens in vitro [25,26]. Finally, we observed that although CD4+ T cells from the child with autosomal-recessive IRF8 deficiency proliferated normally in response to TCR stimulation with CD3 and CD28 (Fig. S5A,C), they poorly secreted effector cytokines IFNγ and IL-17, and to a lesser extent IL10 (Fig. S5B), with a similar pattern detected upon ex vivo stimulation with PMA/ionomycin (data not shown). These results strongly suggest a defect in Th function that may be caused by abnormal T cell differentiation in an environment deficient in antigen-presenting cells.
That autosomal-dominant IRF8 deficiency is associated with a loss of IL-12 producing CD1c+ CD11c+ myeloid DCs suggests that these cells are essential for protective immunity to mycobacteria in humans. The marked reduction in CD1c+ CD11c+ myeloid DCs may be caused by altered ontogeny and maturation of this subset of CD11c+cells, linked to target-specific or global transcriptional effects of the IRF8T80A variant[33]. Despite the fact that mutant whole-blood cells produced normal amounts of IL-12 upon BCG stimulation in vitro, specific impairment of IL12 secretion by CD1c+DCs could contribute to mycobacterial susceptibility. Alternatively, the lack of CD1c+ CD11c+DCs may also contribute to MSMD by other mechanisms, such as impairment of cellular trafficking between draining lymph nodes and infected tissues, or of priming of CD1c-restricted T cells for subsequent activation of infected macrophages. Indeed, CD1c+-restricted T cells specific for a mycobacterial phospholipid antigen have been reported[27,28]. We observed normal levels of CD141+(BDCA3+) DCs -- a subset recently proposed[29–32] to be the functional equivalent of mouse CD8α+ DCs (that are absent in Irf8-deficient mice) in the persons with autosomal-dominant disease. Cells other than CD1c+ DCs may be additionally involved in the pathogenesis of MSMD, and it is possible that IRF8 deficiency impairs effector function of tissue macrophages.
These findings additionally suggest that mutations in hetero-dimerization partners of IRF8 (for example, IRF1 and PU.1) or specific transcriptional targets of such complexes[33] may also impair anti-mycobacterial immunity. Finally, our study illustrates the value of genetic studies in mouse models of infection to identity candidate genes for human primary immunodeficiencies [34].
Supplementary Material
BIBLIOGRAPHY
- 1.Fischer A. Human primary immunodeficiency diseases. Immunity. 2007;27:835–845. doi: 10.1016/j.immuni.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317:617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 4.Geissmann F, Manz MG, Jung S, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flanagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 6.Casanova JL, Jouanguy E, Lamhamedi S, et al. Immunological conditions of children with BCG disseminated infection. Lancet. 1995;346:581. doi: 10.1016/s0140-6736(95)91421-8. [DOI] [PubMed] [Google Scholar]
- 7.Casanova JL, Abel L. Genetic Dissection of immunity to Mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 8.Tamura T, Yanai H, Savitsky D, et al. The IRF family transcription factors in immunity and oncogenesis. Ann Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 9.Crozat K, Guiton R, Guilliams M, et al. Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol Rev. 2010;234:177–198. doi: 10.1111/j.0105-2896.2009.00868.x. [DOI] [PubMed] [Google Scholar]
- 10.Scheller M, Foerster J, Heyworth CM, et al. Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood. 1999;94:3764–3771. [PubMed] [Google Scholar]
- 11.Tamura T, Nagamura-Inoue T, Shmeltzer Z, et al. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 12.Aliberti J, Shulz O, Pennington DJ, et al. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 13.Schiavoni G, Mattei F, Sestili P, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tailor P, Tamura T, Morse HC, 3rd, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol. 2003;170:1131–1135. doi: 10.4049/jimmunol.170.3.1131. [DOI] [PubMed] [Google Scholar]
- 16.Turcotte K, Gauthier S, Malo D, et al. Icsbp1/Irf-8 Is Required For Innate And Adaptive Immune Responses Against Intracellular Pathogens. J Immunol. 2007;179:2467–2476. doi: 10.4049/jimmunol.179.4.2467. [DOI] [PubMed] [Google Scholar]
- 17.Marquis J-F, LaCourse R, Ryan L, et al. Disseminated and rapidly fatal tuberculosis in mice bearing a defective allele at IRF8/ICSBP. J. Immunol. 2009;182:3008–3015. doi: 10.4049/jimmunol.0800680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinh DC, Patel SY, Uzel G, et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115(8):1519–1529. doi: 10.1182/blood-2009-03-208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bigley V, Haniffa M, Doulatov S, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med. 2011 doi: 10.1084/jem.20101459. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtschke T, Lohler J, Kanno W, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 21.Fujii Y, Shimizu T, Kusumoto M, et al. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geissmann F, Gordon S, Hume DA, et al. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010 Jun;10(6):453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merad M, Manz MG, Karsunky H, et al. Langerhans cells renew in the skin throughout life under steady-stae conditions. Nat Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginhoux F, Greter M, Leboeuf M, et al. Fate mapping reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–815. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alter-Koltunoff M, Goren S, Nousbeck J, et al. Innate immunity to intraphagosomal pathogens is mediated by interferon regulatory factor 8 (IRF-8) that stimulates the expression of macrophage-specific Nramp1 through antagonizing repression by c-Myc. J Biol Chem. 2008;283:2724–2733. doi: 10.1074/jbc.M707704200. [DOI] [PubMed] [Google Scholar]
- 26.Fortier A, Doiron K, Saleh M, et al. Restriction of Legionella pneumophila replication in macrophages requires concerted action of the transcriptional regulators Irf1 and Irf8 and nod-like receptors Naip5 and Nlrc4. Infect Immun. 2009;77:4794–4805. doi: 10.1128/IAI.01546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsunaga I, Bhatt A, Young DC, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulrichs T, Moody DB, Grant E, et al. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun. 2003;71:3076–3087. doi: 10.1128/IAI.71.6.3076-3087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jongbloed SL, Kassianos AJ, McDonald KJ, et al. Human CD141+ (BDCA3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulin LF, Salio M, Griessinger E, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells. J. Exp. Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachem A, Guttler S, Hartung E, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crozat K, Guiton R, Contreras V, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to CD8α+ dendritic cells. J Exp Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubosaki A, Lindgren G, Tagami M, Simon C, et al. The combination of gene perturbation assay and ChIP-chip reveals functional direct target genes for IRF8 in THP-1 cells. Mol Immunol. 2010;47(14):2295–2302. doi: 10.1016/j.molimm.2010.05.289. [DOI] [PubMed] [Google Scholar]
- 34.Fortin A, Abel L, Casanova JL, Gros P. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annu Rev Genomics Hum Genet. 2007;8:163–192. doi: 10.1146/annurev.genom.8.080706.092315. [DOI] [PubMed] [Google Scholar]
- 35.Turcotte K, Gauthier S, Tuite A, et al. A mutation in the Icsbp/IRF-8 gene causes susceptibility to infections and a CML-like syndrome in BXH-2 mice. J Exp Med. 2005;201:881–890. doi: 10.1084/jem.20042170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langlais D, Couture C, Balsalobre A, Drouin J. Regulatory network analyses reveal genome-wide potentiation of LIF signaling by glucocorticoids and define an innate cell defense response. PLoS Genet. 2008;4:e1000224. doi: 10.1371/journal.pgen.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eswar N, Webb B, Marti-Renom MA, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5.6) doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Der Spoel D, Lindahl E, Hess B, et al. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 39.Haniffa M, Ginhoux F, Wang XN, et al. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J Exp Med. 2009;206:371–385. doi: 10.1084/jem.20081633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cros J, Cagnard N, Woollard K, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33(3):375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296(5566):261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.