Summary
BMP and Wnt signaling pathways control essential cellular responses through activation of the transcription factors SMAD (BMP) and TCF (Wnt). Here, we show that regeneration of hematopoietic lineages following acute injury depends on the activation of each of these signaling pathways to induce expression of key blood genes. Both SMAD1 and TCF7L2 co-occupy sites with master regulators adjacent to hematopoietic genes. In addition, both SMAD1 and TCF7L2 follow the binding of the predominant lineage regulator during differentiation from multipotent hematopoietic progenitor cells to erythroid cells. Furthermore, induction of the myeloid lineage regulator C/EBPα in erythroid cells shifts binding of SMAD1 to sites newly occupied by C/EBPα, while expression of the erythroid regulator GATA1 directs SMAD1 loss on non-erythroid targets. We conclude that the regenerative response mediated by BMP and Wnt signaling pathways is coupled with the lineage master regulators to control the gene programs defining cellular identity.
Introduction
Cells sense and respond to their cellular environment through signal transduction pathways, which can deliver information to the genome in the form of activated transcription factors. These factors tend to occupy specific genomic regions and associate with different co-activators and chromatin remodeling complexes to direct their response. This occurs by either activating or repressing transcription or by changing the chromatin architecture, thus reforming the accessibility of certain genomic loci (Mosimann et al., 2009; Moustakas and Heldin, 2009). This combination of actions allow for the same signaling pathways to be used in multiple cellular environments eliciting different responses.
The BMP and Wnt signaling pathways are two highly conserved signaling pathways that interact during many developmental processes, ultimately through regulation of transcription via SMAD and TCF/LEF transcription factors (Clevers, 2006; Larsson and Karlsson, 2005; Staal and Luis, 2010). Both pathways participate in the formation of the hematopoietic system during development, but appear to be expendable during adult steady state hematopoiesis (Cheng et al., 2008; Goessling et al., 2009; Jeannet et al., 2008; Koch et al., 2008; Lengerke et al., 2008; McReynolds et al., 2007; Nostro et al., 2008; Singbrant et al., 2010; Tran et al., 2010). In both development and regeneration, hematopoietic stem cells divide and differentiate in response to cell-intrinsic and extrinsic signals to produce all of the hematopoietic lineages. Here we show that the BMP and Wnt signaling pathways are critical for efficient regeneration of the adult hematopoietic system, as they are in development. Additionally, BMP and Wnt have been implicated in differentiation into erythroid and myeloid lineages. Specifically, in culture, BMP treatment can augment erythroid, megakaryocytic, and granulocytic-monocytic output of CD34+ progenitors (Detmer and Walker, 2002; Fuchs et al., 2002; Jeanpierre et al., 2008). Similarly, Wnt3a ligand can regulate the production of erythroid and myeloid cells from ESC and myeloid progenitors from adult HSC (Cheng et al., 2008; Nostro et al., 2008; Staal and Luis, 2010).
The underlying mechanism for BMP and Wnt regulation of regeneration and differentiation resides in the cell-type specific targets of the SMAD and TCF transcription factors, respectively. Based on previous findings, SMAD and TCF proteins can couple with other transcription factors to regulate a small number of cell-specific genes (Clevers, 2006; Massague et al., 2005; Mosimann et al., 2009). Signaling-mediated transcription factors have recently begun to be studied in a genome-wide manner, and these studies have revealed that Smad1 and Tcf7l1/Tcf3 can co-occupy target sites with the Oct4/Nanog/Sox2 transcriptional complex on pluripotency target genes in embryonic stem cells (ESCs) (Chen et al., 2008; Cole et al., 2008) and TCF7L2 can co-localize with CDX2 in colonic cells (Verzi et al., 2010). This led us to consider the possibility that BMP and Wnt signaling factors couple with distinct transcription factors important for lineage identity during hematopoietic regeneration and differentiation.
To determine how SMAD and TCF transcription factors select their targets in distinct lineages during regeneration and differentiation, we explored their genome-wide DNA binding in various hematopoietic environments across multiple species. Initially, co-binding of Smad1 with Gata2 at individual genes in regenerating progenitors was observed. Genome-wide analysis revealed that SMAD1 and TCF7L2 selectively bind in concert with cell-specific master regulators at lineage distinctive genes in erythroid and myeloid cell populations. In addition, the expression of a myeloid master regulator in erythroid cells is sufficient to redirect a fraction of Smad1 binding. During differentiation, the binding of signaling factors shifts from genes of multiple hematopoietic lineages in progenitor cells to genes specific for differentiated cells guided by the dominant lineage factor. Together, these data support a mechanism by which lineage regulators direct SMAD and TCF proteins to tissue-specific enhancers. The selective use of these pathways during regeneration suggests that coordinated binding of SMAD1 and TCF7L2 with lineage-restricted regulators is the underlying mechanism for BMP and Wnt effects during hematopoietic differentiation and regeneration.
Results
Wnt and BMP play essential roles in hematopoietic regeneration
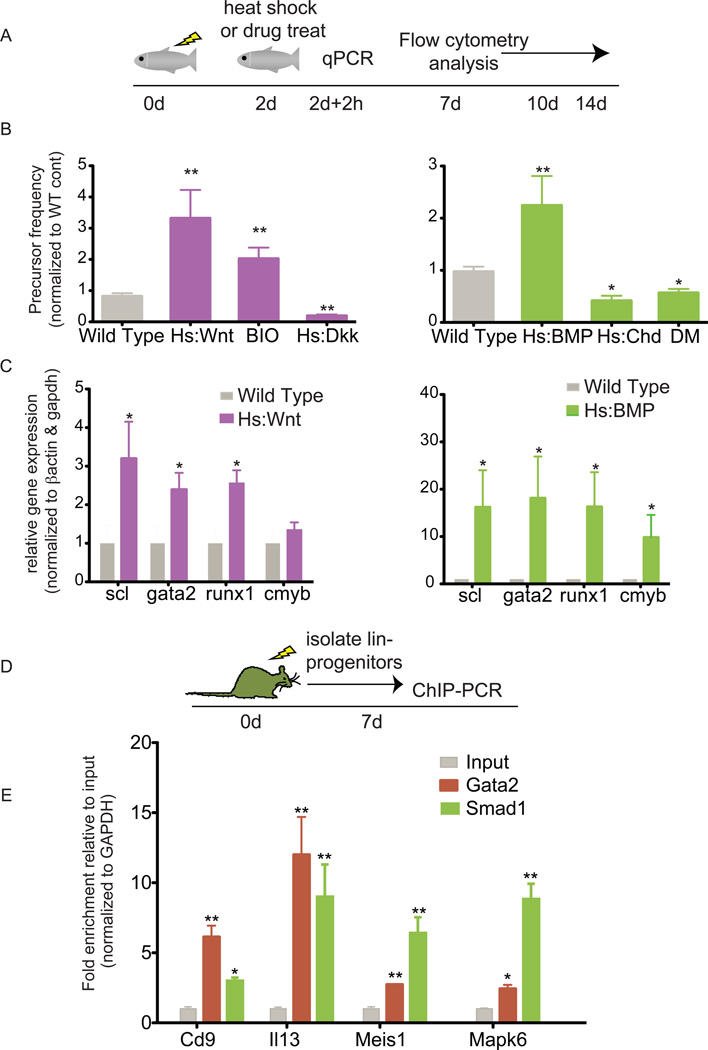
Wnt and BMP signaling are required during hematopoietic development, but it is unclear if either pathway is necessary for adult hematopoietic regeneration. A zebrafish model was used to determine if Wnt or BMP signaling are necessary during this process. Adult zebrafish were sub-lethally irradiated and recovery of whole kidney marrow (WKM) cells was monitored by flow cytometry to identify precursor populations that are the first detectable sign of hematopoietic recovery (Traver et al., 2004) (Figure 1A). Stimulation of the Wnt signaling pathway using a heat shock-inducible Wnt8 ligand (Weidinger et al., 2005), or treatment with the GSK3 chemical inhibitor BIO, resulted in an increase in the frequency of precursors (3- and 2-fold respectively) during regeneration (Figure 1B) (Goessling et al., 2009). In contrast, inhibition of Wnt signaling by heat shock-induced expression of Dkk1 (Stoick-Cooper et al., 2007) led to diminished precursor levels (Figure 1B). Stimulation of the BMP pathway using a heat shock-inducible BMP2 ligand (Rentzsch et al., 2006) enhanced the number of precursors more than 2-fold (Figure 1B), while blocking the BMP pathway using either a heat shock-inducible Chordin (Tucker et al., 2008) or the BMP type I receptor chemical inhibitor dorsomorphin (DM), diminished precursor recovery levels to less than half of the wild-type levels (Figure 1B). Murine competitive bone marrow transplantation experiments were used to assess if the roles for Wnt and BMP in hematopoietic regeneration were conserved and to determine if the effects were cell-intrinsic (Figure S1A). We found that the ex vivo activation of Wnt signaling led to a greater number of animals with multi-lineage engraftment (Figure S1B). In contrast, ex vivo inhibition of BMP signaling abrogated engraftment (Figure S1B). These data suggest that Wnt and BMP have a conserved, cell-autonomous role during adult hematopoietic regeneration.
Figure 1. BMP and Wnt pathways regulate hematopoietic regeneration.
A. Schematic of an irradiation-induced hematopoietic regeneration model. Adult zebrafish are irradiated with 20Gy γ-irradiation at day 0, treated on day 2, and then whole kidney marrow (WKM) are dissected and analyzed by flow cytometry or by QPCR. B. Activation of the Wnt and BMP pathways enhances regeneration in zebrafish. Graphs depicting the relative frequency +/− SEM of precursors in WKM relative to wild type controls following manipulations to the Wnt (left) and BMP (right) pathways. * p<0.05 and **p<0.005, p-values calculated using student’s t-test comparing wild type control treated siblings to treated group. C. Activation of the Wnt (left) and BMP (right) pathways leads to up-regulation of key hematopoietic genes. QPCR graphs of relative gene expression +/− SEM in WKM cells from wild type, Hs:Wnt, or Hs:BMP two days post irradiation following a two hour heat shock induction of Wnt8a or BMP2b transgene expression, respectively. * p<0.05, p-values calculated using student’s t-test comparing wild type control treated siblings to treated group. D–E. Gata2 co-localizes with Smad1 on hematopoietic targets in murine progenitor cells during regeneration. D. Schematic of irradiation model. ChIP for Smad1 and Gata2 was performed on lineage negative progenitors isolated from mice 7 days after a 6.5 Gy irradiation. E. QPCR of whole cell extracts (input), Gata2, and Smad1 ChIP. The bars show relative enrichment +/− SEM compared to input control. * p<0.05 and **p<0.005, p-values calculated using student’s t-test comparing ChIP DNA to input control. See also Figure S1.
The transcriptional effects of BMP and Wnt signaling were evaluated by studying the expression of hematopoietic genes in zebrafish WKM after irradiation following a two hour heat shock induction of BMP2 or Wnt8. Activation of the pathways was confirmed using quantitative PCR for the BMP2 and Wnt8 ligands as well as known downstream targets such as id1 for BMP and cyclind1 for Wnt (Figure S1C). Overexpression of BMP increased the levels of the hematopoietic genes scl, runx1, c-myb, and gata2, while Wnt stimulation increased the expression of scl, runx1, and gata2 (Figure 1C). These data demonstrate that during in vivo regeneration, BMP and Wnt pathways regulate hematopoietic targets at the level of gene expression, which led us to further investigate if the signaling-directed transcription factors directly bind to enhancer elements of blood genes.
We next asked if Smad1, which is a transcription factor activated by BMP signaling, co-occupied hematopoietic genes with the lineage regulator Gata2. In multilineage hematopoietic progenitors, Gata2 is an essential transcription factor that binds to the regulatory elements in genes expressed in progenitors and differentiated lineages (Tsai et al., 1994; Wilson et al., 2010). In order to obtain sufficient cell numbers to perform ChIP analysis, a mouse irradiation injury model was used (Hooper et al., 2009). Lineage-negative progenitors were isolated from mice seven days after a sublethal irradiation, and then ChIP-PCR was performed for known targets of Gata2 (Figure 1D) (Wilson et al., 2010). Smad1 and Gata2 co-occupied hematopoietic genes including Cd9, Il13, Meis1, and Mapk6 (Figure 1E). Together, these results indicate that BMP and Wnt are required for regeneration and act at least in part by modulating genes bound by the lineage regulator Gata2.
TCF7L2 and SMAD1 transcription factors co-occupy genomic sites with GATA factors in an erythroid environment
We next asked if the transcription factors activated by Wnt and BMP signaling co-bind with blood-specific lineage regulators genome-wide by performing ChIP-seq for TCF7L2/TCF4 (Wnt pathway) and SMAD1 (BMP pathway) as well as GATA1 and GATA2. ChIP-seq was performed in K562 cells, which are erythroleukemia cells that express GATA1 and GATA2, essential transcription factors for erythroid and progenitor cells, respectively (Cantor, 2005; Tsai et al., 1994; Tsiftsoglou et al., 2009). Cells were treated with BIO to activate the Wnt pathway or BMP4 to activate the BMP pathway. Both TCF7L2 and SMAD1 showed substantial overlap with GATA1 and GATA2 at individual genes and genome wide (Figure 2A and 2B). To confirm the specificity of the analysis, TCF7L2 and SMAD1 binding at known targets was analyzed (Figure S2A) and found that inhibition of BMP signaling resulted in considerable loss of SMAD1 binding across the genome (Figure S2B).
Figure 2. SMAD1 and TCF7L2 co-occupy the genome with key regulators of the erythroid lineage.
A. Gene track of the GATA1 locus showing TCF7L2 (purple), GATA1 (red), GATA2 (orange), and SMAD1 (green) binding of specific genomic regions along the x-axis and the total number of reads per million on the y-axis. B. SMAD1 and TCF7L2 co-occupy genomic regions with GATA1 and GATA2. Region plots representing the distribution of GATA1 and GATA2 bound regions −2.5 to +2.5 kb relative to all TCF7L2 or SMAD1 bound regions in K562 cells. C. QPCR of whole cell extracts (input), SMAD1 and control mouse IgG, sequential ChIP for GATA2 and control rabbit IgG on the SMAD1 ChIP. The bars show relative enrichment +/− SEM compared to input control. * p<0.08 and **p<0.04, ***p<0.0005, p-values calculated using student’s t-test comparing SMAD1 ChIP to input and SMAD1 ChIP to SMAD1-GATA2 sequential ChIP. D. Co-localization of SMAD1 and TCF7L2 are specific to lineage regulators. Heat map depicting the relative level of co-localization of indicated factors, in K562 cells together with open chromatin data in this cell line. E. E2F4 and CTCF binding is not associated with TCF7L2 or SMAD1. The distance from the center of each TCF7L2 site (left) and SMAD1 site (right) to the center of the nearest site bound by the indicated transcription factor was determined. These distances were grouped into bins (x-axis). The sum of bound sites in each bin is shown (y-axis). See also Figure S2.
If the main targets for TCF7L2 and SMAD1 are the blood genes regulated by GATA factors, then we should see enrichment for red blood cell genes in their targets. Functional classification of the genes occupied by TCF7L2 or SMAD1 in K562 erythroleukemia cells revealed enrichment for genes highly associated with red blood cell development(Figure S2C–E). If TCF7L2 and SMAD1 bind to the same sites as GATA1 and GATA2, then there should be enrichment for GATA motifs in the sites bound by TCF7L2 or SMAD1. As expected, the most prominent motif in regions bound by TCF7L2 and SMAD1 was the GATA motif (Figure S2F). Scanning for GATA, SMAD, and TCF motifs across the regions bound by SMAD1, TCF7L2, GATA1 or GATA2 demonstrated enrichment of all three motifs in all of the bound sites (Figure S2G). Furthermore Wnt and BMP stimulation did not significantly affect the binding of GATA factors (Figure S2H). If GATA factors direct the binding of signaling factors, then the expectation would be to find them interacting with the same genomic sites at the same time. ChIP for SMAD1 was performed followed by ChIP for GATA2, and PCR showed that SMAD1 and GATA2 simultaneously occupied key erythroid genes (Fig 2C). These results are consistent with co-occupancy of TCF7L2 and SMAD1 with GATA1 and GATA2 in K562 cells and demonstrated that Wnt and BMP signaling were directed to genes occupied by GATA factors that specified the red blood cell state.
We next asked if TCF7L2 or SMAD1 preferentially occupied the genome with GATA1 and GATA2 in K652 cells. Available data were analyzed for other transcription factors in K562 cells (Birney et al., 2007; Raney et al., 2011; Rosenbloom et al., 2010). Both TCF7L2 and SMAD1 tend to bind sites with GATA1 and GATA2 but do not tend to occupy sites with the other tested transcription factors despite previous reports that SMAD1 could interact with several of these factors in other tissue types (Chen et al., 2002; Kurisaki et al., 2003) (Fig 2D). In addition, the distance from TCF7L2 or SMAD1 sites to the nearest binding site for GATA1, GATA2, E2F4, and CTCF was calculated. The majority of TCF or SMAD sites were occupied by the GATA factors, but were distant from E2F4 and CTCF sites (Figure 2E). These data indicate that the genome-wide co-occupancy of TCF7L2 and SMAD1 with GATA1 and GATA2 is specific.
Co-occupied regions encompass enhancers
Analysis of TCF7L2 and SMAD1 localization with multiple transcription factors in K562 cells, revealed that each of the transcription factors examined occupied “open” chromatin sites as defined by FAIRE-Seq and DNAse-Seq, which measure DNA accessibility (Song et al., 2011), indicating TCF7L2 and SMAD1 do not indiscriminately bind to all open and available DNA sites (Figure 2D). This led us to investigate the class of regulatory elements occupied by BMP and Wnt signaling factors.
Transcriptional regulatory elements fall into well-defined groups that can be defined by position relative to gene, chromatin modification state and occupancy by known regulators. In order to identify the class of regulatory elements Wnt and BMP factors co-occupy with master regulators, the enriched regions were mapped to positions relative to RefSeq genes. To assess differences in regions co-bound by signaling and lineage regulators vs. those bound by a lineage regulator alone, regulatory elements in regions bound by GATA factors and TCF7L2 or SMAD1 were examined and compared to those bound by GATA factors alone. The majority of regions for all groups mapped to introns and intergenic regions, similar to many enhancer elements (Heintzman and Ren, 2009), in contrast to E2F4, which occupies sites adjacent to promoter regions (Figure 3A). This pattern suggests TCF7L2 and SMAD1 co-occupy distal regulatory elements with GATA1 and GATA2 master regulators.
Figure 3. Signaling factors cooperate with lineage regulators at distal enhancers.
A. TCF7L2 and SMAD1 regions co-localize with GATA factors in intronic and intergenic regions. The groups of enriched regions occupied by GATA factors in K562 cells were divided into those occupied by GATA only, TCF7L2 and GATA, or SMAD1 and GATA. E2F4 is shown as a control. Each region was mapped to their closest RefSeq gene: distal promoter (blue), proximal promoter (red), exons (green), introns (purple) and intergenic regions (light blue) B. TCF7L2 and SMAD1 regions that co-localize with GATA factors occupy mainly enhancer regions. Composite H3K4me1-BIO (purple) and H3K4me1-BMP4 (green) enrichment profile for TCF7L2 and GATA co-bound regions, SMAD1 and GATA co-bound regions or regions occupied only by GATA in K562 cells. C. Gene track of the LMO2 locus. A graph of the LMO2 reporter indicating the enhancer and promoter regions included in the construct is shown below. D. SMAD1 and TCF7L2 co-operate with GATA2 and enhance transcription of target genes. Graph depicting β-galactosidase activity of the reporter +/− SEM following overexpression of the transcription factors listed under each bar. *p<0.06, **p<0.01, p-values calculated using student’s t-test comparing mock transfected controls to GATA2 alone and SMAD1/GATA2 or TCF7L2/GATA2 co-transfections to GATA2 alone. E. Wnt and BMP signaling enhance p300 recruitment to blood-specific targets. ChIP-PCR graphs showing p300 occupancy after activation or inhibition of the Wnt (left) and the BMP (right) pathways. *p<0.06, **p<0.01, p-values calculated using student’s t-test comparing inhibitor-treated samples to activator-treated samples.
Mammalian enhancers are associated with the histone modification H3K4me1 (Heintzman et al., 2009; Heintzman et al., 2007; Rada-Iglesias et al., 2011; Visel et al., 2009). Therefore we asked whether the distinct GATA regions showed differences in these modifications. While each group was marked by H3K4me1 surrounding the GATA-bound regions, those sites where TCF7L2 or SMAD1 co-localize with GATA factors showed a greater enrichment (Figure 3B). To determine if these enhancers tend to be associated with actively transcribed genes, the transcriptional activity of genes within 5 kb of the enriched regions was classified into distinct states of active, poised and silent. Active genes were defined as the Refseq genes marked by the histone modifications H3K4me3 and H3K36me3. A higher proportion of regions were enriched at active genes (51–54%) compared to the proportion of all active genes (35%) (Figure S3). Thus, the co-occupied regions of TCF7L2, SMAD1 and GATA factors are predominantly occupying enhancers of actively transcribed genes. Together, these observations predict that the expression level of GATA target genes would be responsive to perturbation of Wnt and BMP signaling.
TCF7L2 and SMAD1 enhance transcriptional activation mediated by GATA2
TCF7L2 and SMAD1 co-occupy enhancers of active, cell type-specific genes with GATA factors. To assess the consequences of this co-occupancy on transcriptional output, the effects of each transcription factor alone or in combination were examined on the expression of the hematopoietic gene LMO2, which is bound by TCF7L2, SMAD1, GATA1, and GATA2 72kb upstream of the transcriptional start site in K562 cells (Figure 3C)(Landry et al., 2009). Induction of GATA2 alone was sufficient to increase reporter expression, while neither SMAD1 nor TCF7L2 alone had any effect (Figure 3D). In contrast, induction of either SMAD1 or TCF7L2 in the presence of GATA2 greatly enhanced reporter activity, indicating that the signaling factors affect transcription of blood-specific enhancer elements in combination with a lineage regulator (Figure 3D).
The observation that an increase in histone modifications is associated with active enhancers bound by TCF7L2, SMAD1, and GATA factors led to the hypothesis that binding of signaling factors could be influencing the chromatin state at these elements to affect transcriptional output. This could occur through recruitment of the histone acetyltransferase p300, with which both TCF/β-catenin and SMAD are known to interact (Hecht and Stemmler, 2003; Nakashima et al., 1999; Pearson et al., 1999). If recruitment of p300 was dependent on the activity of the Wnt and BMP signaling pathways, then perturbations to either Wnt or BMP signaling should affect the localization of p300 to the enhancers of blood-specific genes. Activation of either the Wnt or the BMP pathways in K562 cells did result in an increase of p300 occupancy by ChIP-PCR, whereas inhibition of the pathways diminished p300 occupancy at the enhancer elements of blood genes (Figure 3E). We conclude that TCF7L2 and SMAD1 co-operate with lineage regulators to enhance the transcription of their target genes through co-occupancy of enhancers that are activated through recruitment of p300.
TCF7L2 and SMAD1 transcription factors co-occupy genomic sites with C/EBPα in a myeloid environment
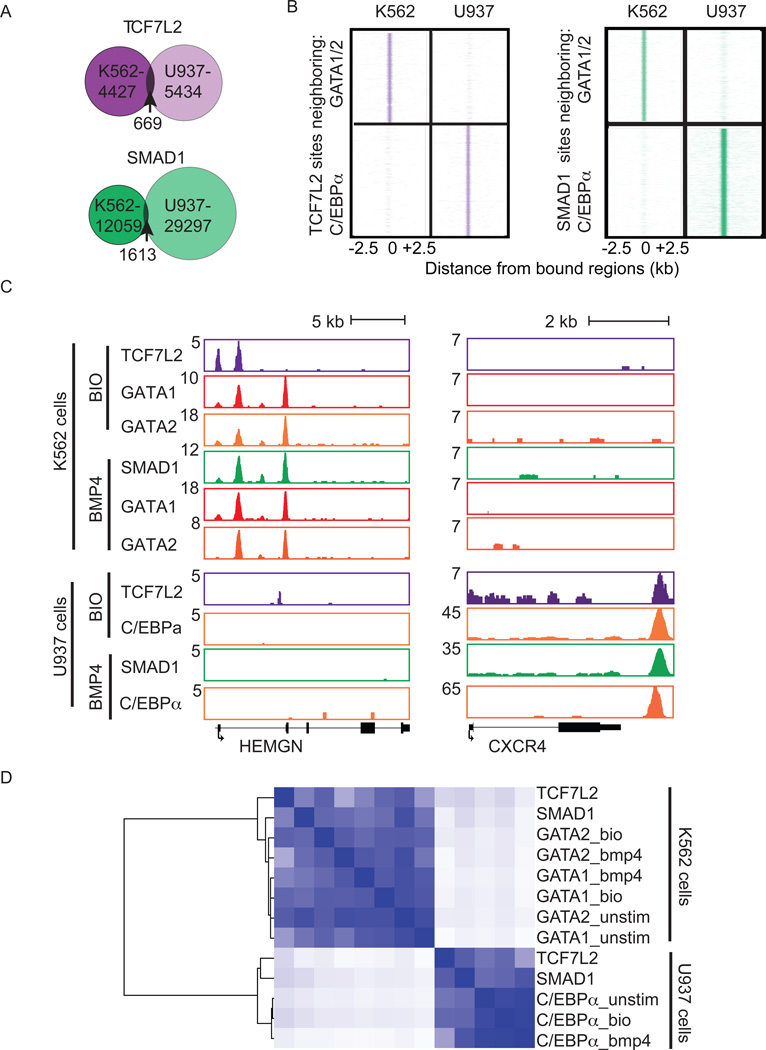
We next investigated if TCF7L2 and SMAD1 occupied the genome with a different master regulator in another hematopoietic lineage. ChIP-Seq was performed for TCF7L2, SMAD1 and C/EBPα, a known master regulator of the myeloid lineage, in the U937 monocytic leukemia cell line. TCF7L2 and SMAD1 tend to occupy different sites in erythroid (K562) and myeloid (U937) lineages, with only 15% of sites in common (Figure 4A). Furthermore the sites occupied by TCF7L2 and SMAD1 tend to be the sites occupied by C/EBPα in U937 cells and by GATA in K562 cells (Figure 4B and 4C). In the erythroid environment, TCF7L2, SMAD1, GATA1, and GATA2 were bound within erythroid genes such as HEMGN, but those sites were not bound by TCF7L2, SMAD1, or C/EBPα in the myeloid environment (Figure 4B, 4C, S4A and S4B). Similarly, genes with a stronger association with myeloid cells such as CXCR4 were bound by TCF7L2, SMAD1, and C/EBPα in the myeloid environments, but not bound by TCF7L2, SMAD1, GATA1, or GATA2 in the erythroid environment (Figure 4B, 4C, S4A and S4B). Motif scanning of the sequences around TCF7L2, SMAD1 or GATA bound sites in K562 showed enrichment for GATA, SMAD, and TCF motifs and absence of C/EBP motifs, whereas in the myeloid cells C/EBP motifs were present and GATA motifs absent (Figure S4C). Treatment of U937 cells with BIO or BMP4 had little influence on C/EBPα binding showing activation of the Wnt or BMP signaling pathways does not affect the genomic localization of the myeloid regulator (Figure S4D).
Figure 4. TCF7L2 and SMAD1 co-occupy genomic regions with cell-type-specific lineage regulators.
A. Venn diagrams depicting the overlap of regions bound in K562 and U937 cells for TCF7L2 (top) and SMAD1 (bottom). Numbers of regions bound by each factor in each cell line or in the overlap of both are shown. B. Region plots comparing the enriched regions of TCF7L2 and SMAD1 in K562 and U937 cells compared to GATA1 and GATA2 (top) or C/EBPα (bottom) regions. C. Gene tracks of HEMGN (left) and CXCR4 (right) showing differential binding of TCF7L2, SMAD1, GATA1, GATA2 and C/EBPα in K562 cells (top) and U937 cells (bottom). D. Heat map depicting the co-localization of GATA1, GATA2, TCF7L2, and SMAD1 in K562 cells and C/EBPα, TCF7L2, and SMAD1 in U937 cells. See also Figure S4.
Analysis of bound regions among all the transcription factors analyzed showed a strong cell type clustering (Figure 4D). TCF7L2 and SMAD1 bound sites in K562 show a stronger correlation with GATA1 and GATA2 occupied regions, than TCF7L2 and SMAD1 bound sites in U937. These data suggest that TCF7L2 and SMAD1 show cell lineage specific binding at sites co-occupied by master regulators.
C/EBPα expression re-directs SMAD1 binding in erythroid cells
If lineage-specific transcription factors direct the Wnt and BMP regulators to regulatory elements in distinct lineages, then expression of a master regulator of the myeloid lineage (e.g. C/EBPα) in an erythroid cell-type should be capable of re-directing some of the signaling factors to novel sites occupied by the myeloid regulator. In order to test this hypothesis, an estrogeninducible C/EBPα expressed in K562 cells was used (D'Alo et al., 2003). Upon estradiol treatment, the C/EBPα estrogen receptor fusion protein translocates into the nucleus. SMAD1 and C/EBPα binding was determined by ChIP-seq in these cells following a 24-hour estradiol induction and a 2-hour BMP4 stimulation (Figure 5A). Expression of C/EBPα directed a fraction of SMAD1 to new sites occupied by C/EBPα (Figure 5B). SMAD1 binding was retained at GATA targets, such as SLC6A9, in C/EBPα-expressing K562 cells, while occupying new sites with C/EBPα exclusive of erythroid regulators (Figure 5C). An example of a newly occupied site lies near two genes, ALAS2 and APEX2. ALAS2 is expressed in erythroid cells (Sadlon et al., 1999), while APEX2 is important in white blood cells (Ide et al., 2004). The binding of C/EBPα and SMAD1 at this position may result in repression of ALAS2 in erythroid cells or activation of APEX2 in white blood cells.
Figure 5. C/EBPα expression re-positions SMAD1 binding in K562 cells.
A. Schematic of C/EBPα-ER K562 experimental model. B. The percentage of SMAD1 sites in K562 cells that are co-occupied by C/EBPα (y-axis) is shown for K562 cells (no C/EBPα) and those induced by C/EBPα (+C/EBPα). The top 1000 SMAD1 binding sites in each condition were used for this calculation. C. Gene tracks of SLC6A9 (left) and ALAS2/APEX2 (right) showing differential binding of GATA1, GATA2 and SMAD1 in K562 cells (top) SMAD1 and C/EBPα in K562 cells expressing C/EBPα (middle) and U937 cells (bottom).
Gata1 expression re-directs SMAD1 binding during differentiation
Ectopic expression of C/EBPα is sufficient to direct SMAD1 to some new targets, but does not induce significant loss of SMAD1 binding on other targets. We next asked whether forced expression of an erythroid master regulator (Gata1) in an erythroblast progenitor cell line could restrict Smad1 binding to only erythroid targets. To address this question, the estrogeninducible Gata1-null erythroblast cell line (G1ER) was used, which was derived from targeted disruption of Gata1 in embryonic stem cells. Upon estradiol treatment, the Gata1 estrogen receptor fusion protein translocates into the nucleus and induces the red blood cell differentiation program (Figure 6A) (Cheng et al., 2009; Tsang et al., 1997; Yu et al., 2009). We identified the genome-wide binding of Gata2 and Smad1 in Gata1-deficient (G1E) and Gata1 and Smad1 in Gata1-induced (G1ER) cells following BMP4 stimulation. SMAD and GATA motifs were identified in all samples (Figure S6A). In G1E cells, Smad1 and Gata2 co-occupied genes of multiple hematopoietic lineages (Figure 6B, 6C, S6B, and S6C). During erythroid differentiation Gata1 replaces Gata2 binding at erythroid genes (Bresnick et al., 2010; Grass et al., 2003). After Gata1 induction, Smad1 binding became more restricted to erythroid genes, and binding to genes expressed in the other lineages was diminished (Figure 6B, 6C, S6B, and S6C). Together the data reveal that induction of lineage-specific regulators alter Smad1 genomic occupancy.
Figure 6. Smad1 localization is directed by Gata1.
A. Schematic of the G1E, G1ER experiment. B. Gene tracks of Flt3 (top) and Alas2 (bottom) showing differential binding of Gata2 and Smad1 in G1E Gata1-null cells (Proerythroblast), and Gata1 and Smad1 in G1ER erythroid cells (Differentiating). C. Overexpression of Gata1 re-defines the targets of Smad1. ChIP-seq region plots representing the distribution of regions bound by Smad1 and Gata2 in G1E, and Gata1 and Smad1 in G1ER cells −2.5 and +2.5 kb relative to all Smad1 bound sites in G1E and G1ER combined.
TCF7L2 and SMAD1 co-occupancy with master regulators occurs in primary hematopoietic progenitors and changes during erythropoiesis
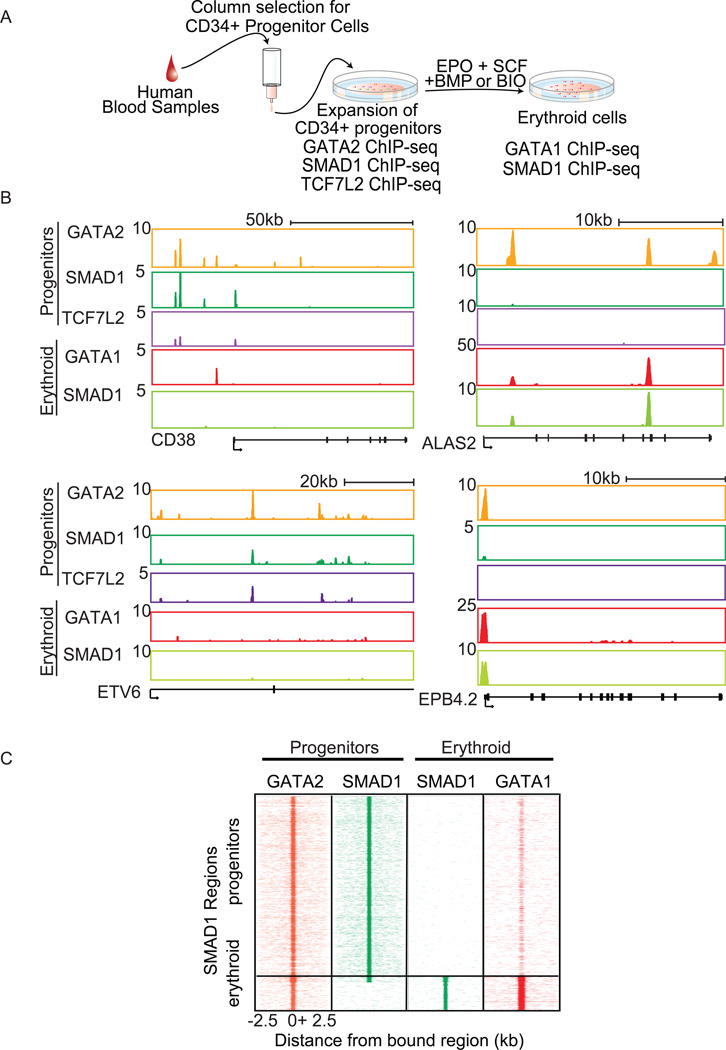
The shifts in SMAD1 occupancy following forced expression of C/EBPα or Gata1 in erythroid environments suggest that during regeneration and differentiation, when there are dynamic changes in the lineage regulators present, signaling factor occupancy will also change according to the cell state. To model a normal differentiation process, analogous to what occurs during regeneration, we examined TCF7L2 and SMAD1 genomic occupancy in primary human progenitor cells following in vitro expansion and differentiation. ChIP-seq was performed for GATA2, TCF7L2, and SMAD1 in mobilized peripheral blood CD34+ progenitor cells (Pro) (Figure 7A and S7A). GATA2 co-occupied more than 75% of TCF7L2 or SMAD1 enriched genes in these hematopoietic progenitor cells (Figure 7B, 7C, S7B, S7C, and S7D). In addition to a GATA motif, ETS and RUNX motifs were also enriched in GATA2, SMAD, and TCF bound sequences (Figure S7E). This finding is consistent with recent genome-wide data from murine hematopoietic progenitors that indicate Gata2 co-occupies sites with multiple regulators including Runx1 and ETS transcription factors Erg and Fli1 near genes important for multipotent stem cells (Wilson et al., 2010). Together with the binding data, these results imply that TCF7L2 and SMAD1 are likely co-binding with the entire progenitor transcriptional complex.
Figure 7. Binding of signaling factors changes during differentiation.
A. Schematic of CD34+ experiment. B. Gene tracks of CD38, ETV6, ALAS2, and EPB4.2 showing differential binding of GATA2, SMAD1 and TCF7L2 in undifferentiated CD34+ progenitors, and GATA1 and SMAD1 in differentiated erythroblasts. C. SMAD1 binding becomes restricted to mainly erythroid genes after differentiation of CD34+ hematopoietic cells towards erythrocytes. ChIP-seq region plots representing the distribution of SMAD1 occupied regions in CD34 progenitors (pro) and in vitro differentiated erythroblasts (ery). GATA2 and SMAD1 in CD34+ progenitors and SMAD1 and GATA1 in erythroid differentiated CD34 cells −2.5 to +2.5 kb relative to all SMAD1 bound regions in CD34 progenitors and differentiated erythroblasts. See also Figure S7.
To determine how binding changes during differentiation, ChIP-Seq for GATA1 and SMAD1 in erythroblast derived from CD34+ progenitors (CD34ery) was performed (Figure 7A and S7A). SMAD1 co-bound with GATA1 in CD34-derived erythroblasts (Figure 7B and 7C). The genes bound by SMAD1 were fewer in the erythroid cells compared to the progenitor cells (2683 vs.8094). This change in SMAD1 occupancy suggested refinement of the binding sites from a multilineage to a fully differentiated state (Figure 7B and 7C). The bound genes were enriched for those that are characteristic of the differentiation stage (Figure S7D). In addition to the loss of many bound regions, further binding was acquired at erythroid genes (Figure 7B, 7C and S7D). These data confirm that co-binding of BMP and Wnt regulators with lineage transcription factors occurs in primary hematopoietic cells in genes that regulate all hematopoietic lineages and shifts upon directed differentiation towards erythroid cells to erythroid-specific genes.
To ascertain if genes bound by BMP and Wnt transcription factors were regulated by BMP and Wnt signaling, genome-wide expression analysis was performed, on CD34+ cells with or without stimulation by BMP4 or BIO, respectively. The expression in unstimulated and stimulated cells was compared two hours after the shift to erythroid differentiation conditions. Differentially expressed genes in BMP4 treated groups compared to untreated controls were enriched for genes bound by SMAD1 (32%), and those differentially expressed in BIO treated samples were enriched for those bound by TCF7L2 (31%) (Table S6). These data suggest that BMP and Wnt signaling coordinate with differentiation signals to alter gene expression via direct binding to common target genes with lineage master regulators.
Discussion
Here we provide evidence that BMP and Wnt pathways play a dynamic role in hematopoietic regeneration through co-occupancy of regulatory elements with lineage regulators at cell-type specific genes for each lineage. The co-occupancy of Smad1 and Gata2 was observed on blood targets in hematopoietic progenitors during in vivo regeneration. Lineage-restricted co-occupancy of TCF7L2 or SMAD1 with GATA1 and GATA2 in erythroid cells, C/EBPα in myeloid cells, and GATA2 in progenitors occur genome-wide. This binding is selective for cell-specific enhancers bound by master regulators. BMP and Wnt signaling cooperate with lineage regulators to enhance transcription of cell-type specific target genes. Lastly, ectopic expression of a lineage transcription factor was sufficient to direct the genomic localization of signaling-specific factors.
Master regulators direct the site selection of signaling transcription factors in every step during the differentiation process. Recent genome wide studies have shown that signaling pathway transcription factors localize in binding sites adjacent to embryonic stem cell master regulators (Chen et al., 2008; Cole et al., 2008; Young, 2011). Our data show that this mechanism occurs in many cell types, and establishes an order by which the lineage regulators recruit the signaling factors to sites of active genes of biological relevance throughout the whole genome. Alterations of the expression and binding of hematopoietic lineage regulators to target genes during regeneration or differentiation can dictate the binding of signaling factors.
Signaling factors selectively co-localize with master regulators, but our studies hint that other factors may help fine-tune signaling factor binding. For example, the transcriptional status of a gene can modify SMAD1 binding. In CD34 progenitor cells, SMAD1 co-binds with GATA2 on genes expressed in progenitors, but is mostly absent from erythroid genes that are not expressed. In erythroid cells, SMAD1 is able to bind these same targets with GATA1, which helps activate these genes (Bresnick et al., 2010; Grass et al., 2003). Additionally, It has been suggested that GATA2 works in different complexes to target progenitor vs. erythroid genes (Wilson et al., 2010), and GATA2 is expressed at different levels in these two cell populations. We speculate that it is not only the function of an individual master regulator, but also the combinations and the levels of each lineage regulator in a cell along with the transcriptional state of the target that help dictate the genomic location of signaling factors.
Here we provide a model for how this mechanism could be utilized to help orchestrate hematopoietic differentiation during a stress response. During hematopoietic regeneration when these pathways are required, the activation of BMP and Wnt signaling results in the co-localization of SMAD and TCF with the master regulators on genes defining progenitor cell fate. As regeneration continues and progenitor cells begin to differentiate, different master regulators activate the cell-specific genes of more mature hematopoietic lineages and again redefine the binding of signaling transcription factors. Co-localization of lineage and signaling factors has as a result the co-localization of signaling factors themselves. This fact may explain some of the synergistic effects observed between many signaling pathways (Schier and Talbot, 2005). As the BMP and Wnt pathways appear to have a selective function during regeneration, throughout this stress response, coupling transcriptional regulation to the transcription factors expressed highly in a cell lineage explains how BMP and Wnt signaling pathways can have cell-context dependent effects.
Regeneration is a process of tissue self-renewal. Our data imply that an underlying mechanism of self-renewal is the control of the entire hematopoietic program through the collaboration of master and signaling transcription factors, which is similar to the mechanism by which core self-renewal factors in embryonic stem cells perform key roles in embryonic stem cell fate. Our observation that the BMP and Wnt signaling transcription factors are associated with master regulators of multiple hematopoietic cell types provide insight into the effects of BMP and Wnt signaling that occur during development, regeneration, and disease.
Experimental Procedures
Zebrafish Irradiation-induced Regeneration
Zebrafish were maintained as described (Westerfield, 2000). Irradiation-induced regeneration assays were performed as previously described (Burns et al., 2005; Traver et al., 2004). For heat shock treatments, small fish tanks were placed at 37°C for 16 hours. For small molecule treatments, fish were soaked for 16 hours in fish water containing drug dissolved in DMSO, 2µM BIO or 10µM DM. On days 2, 7, and 14 post irradiation, whole kidney marrow cells were analyzed on an LSRII for FSC/SSC parameters. See also Extended Experimental Procedures.
Cell line culture conditions and stimulation
K562 and U937 were maintained in IMDM and RPMI medium, respectively, supplemented with fetal calf serum, glutamine, and penicillin/streptomycin. Culture conditions for CEBPa-K562, G1E, G1ER, and human hematopoietic CD34+ cells as well as differentiation media and stimulation conditions are described in Extended Experimental Procedures.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation experiments were performed as previously described (Lee et al., 2006) with slight modifications, which are described in Extended Experimental Procedures. A summary of the bound regions and bound genes determined for all ChIP-seq data is contained within Supplementary Table 3,Supplementary Table 4,Supplementary Table 5. For ChIP-seq experiments the following antibodies were used: Smad1 (Santa Cruz sc7965), TCF7L2 (TCF4 Santa Cruz sc8631), C/EBPα (Santa Cruz sc9314), Gata1 (Santa Cruz sc265), Gata2 (Santa Cruz sc9008), p300 (Santa Cruz sc585), and H3K4me1 (Abcam ab8895). Samples were prepared for sequencing using Illumina Genomic DNA kit or TruSeq according to the manufacturers instructions. Also see Extended Experimental Procedures for details on ChIP and ChIP-seq sample preparation and analysis.
ChIP-PCR analysis of lineage negative bone marrow cells
ChIP in lineage negative cells isolated from mouse bone marrow was performed as described above. Primers were designed for known Gata2 targets. QPCR reactions were done with iQ SYBR-GREEN Supermix (BIORAD 170–8880), and a CFX-384 Real-Time PCR Detection System (BIORAD) thermal cycler was used. Expression of each gene was normalized to Gapdh and relative levels were calculated using the ΔΔCt method (Applied Biosystems). Quantities are expressed as fold change compared to input controls. Primers are listed in Supplementary Table 2.
Sequential Chromatin immunoprecipitation
ChIP were performed and immunoprecipitated DNA fragments were eluted from the beads by addition of 55µl 10mM DTT and incubated at 37°C for 30 min. The supernatant was transferred to a new tube and the material was diluted 30X with sonication buffer and served as the starting material for the second ChIP. ChIP-PCR was performed as described above. See also Extended Experimental Procedures.
Motif counting
The genomic sequence +/−2.5 kb from the center of each enriched region in the dataset indicated was downloaded from the UCSC website with repeats masked with “N”. A window of 250bp was shifted across each sequence at 50bp intervals, and the number of occurrences of the complete motif sequence or its reverse complement within the window was counted. An averaged motif occurrence in each scanning window was plotted across the +/−2.5kb regions.
Heatmap analysis of ChIP-seq occupancy
ChIP-seq enrichment for the indicated factor or modification was determined in 100bp bins (enrichment in the bin as counts per million) centered on the enriched region of a given ChIP-seq dataset. Java Treeview (www.jtreeview.sourceforge.net) was used to visualize the data and generate figures shown in this manuscript.
Distance from TCF7L2 or SMAD1 enriched regions to nearest TF bound region
The distance (in bp) from each TCF7L2 or SMAD1 site in K562 cells to the nearest site of each TF was calculated. The distance from the boundary of each TCF7L2 or SMAD1 enriched region to the boundary of the nearest site bound by the indicated transcription factor was determined. These distances were grouped into bins (x-axis), with the x-axis value indicating the upper limit of each bin. The sum of regions in each bin is shown (y-axis).
Assignment to regulatory regions in K562 Cells
Each enriched region was uniquely assigned into a category of exon, intron, proximal promoter (from 1KB upstream to TSS), distal promoter (from 10KB upstream to 1KB upstream of TSS), and intergenic region (more than 10KB upstream away from body of the gene) according to MySQL access to the UCSC hg18 refseq table. When an enriched region could be mapped to more than 1 regulatory region category because of overlapping genes, it was classified into a regulatory region class in preferential order of exon, intron, proximal promoter, distal promoter, and intergenic region.
Genome-wide expression analysis
Affymetrix U133plus2.0 microarrays were used to assess gene expression changes in CD34+ cells with or without Wnt or BMP pathway stimulation. Arrays were done on control, 0.5µM BIO, or 25ng/ml rhBMP4 treated cells two hours after a shift to erythroid differentiation media. See also Extended Experimental Procedures.
Reporter assays
K562 cells were transfected using AMAXA nucleofector according to the manufacturer instructions. β-galactosidase activity was measured with the Galacto-Star kit (Applied Biosystems). See also Extended Experimental procedures.
Highlights.
* BMP and Wnt pathways regulate hematopoietic regeneration
* SMAD1 and TCF7L2 co-occupy genomic regions with GATA factors in erythroid cells
* Regions co-occupied by signaling factors and lineage regulators are enhancers
* Expressing myeloid lineage regulator C/EBP in erythroid cells redirects SMAD binding
Supplementary Material
Acknowledgments
We would like to acknowledge A. Mullen and D. Orlando for helpful discussions and sharing critical insights prior to publication and G.Frampton for analytical advice and development of the ChIP-seq analysis platform. We thank F. Rentzsch and M. Hammerschmidt for the Hs:BMP zebrafish line, J. Tucker and M. Mullins for the Hs:Chordin zebrafish line, and T. Schlaeger for the GATA2 expression plasmid. We are grateful to X. Bai and R. White for critical reviews of the manuscript and O. Tamplin for computational advice. The Whitehead Genome Technology Core was instrumental in timely data production and analysis support, especially S. Gupta. Microarray studies were performed by the Molecular Genetics Core Facility at Children's Hospital Boston supported by NIH-P50-NS40828, and NIH-P30-HD18655.
This work was supported by NIH #5PO1HL32262-29 and #5R01HL048801-18 (to L.I.Z), NIH 1R01DK080040-01 (to R.F.P.), R01-HG002668 (to R.A.Y.), NIH K01 DK085270-02 (to T.V.B.), NIH U01 HL100001, NIH 1 RC2HL102815 and ROFAR (to G.Q.D.), NIH T32HL007623 (to G.C.H.), HHMI (to LI.Z. and G.Q.D.), and EMBO fellowship and Jane Coffin Childs Memorial Fund (to E.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Microarray (GSE26351) and ChIP-seq (Superseries-GSE29196, datasets-GSE29193, GSE29194, GSE29195) data were deposited on GEO (www.ncbi.nlm.nih.gov/geo/) under the accession numbers indicated.
Conflicts of interest: L.I.Z. is a founder and stock holder of Fate, Inc. and a scientific advisor for Stemgent. G.Q.D. is a member of the Scientific Advisory Boards of MPM Capital, Inc., Epizyme, Inc., and iPierian, Inc.
References and Notes
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB. GATA transcription factors in hematologic disease. Int J Hematol. 2005;81:378–384. doi: 10.1532/ijh97.04180. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng X, Huber TL, Chen VC, Gadue P, Keller GM. Numb mediates the interaction between Wnt and Notch to modulate primitive erythropoietic specification from the hemangioblast. Development. 2008;135:3447–3458. doi: 10.1242/dev.025916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alo F, Johansen LM, Nelson EA, Radomska HS, Evans EK, Zhang P, Nerlov C, Tenen DG. The amino terminal and E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood. 2003;102:3163–3171. doi: 10.1182/blood-2003-02-0479. [DOI] [PubMed] [Google Scholar]
- Detmer K, Walker AN. Bone morphogenetic proteins act synergistically with haematopoietic cytokines in the differentiation of haematopoietic progenitors. Cytokine. 2002;17:36–42. doi: 10.1006/cyto.2001.0984. [DOI] [PubMed] [Google Scholar]
- Fuchs O, Simakova O, Klener P, Cmejlova J, Zivny J, Zavadil J, Stopka T. Inhibition of Smad5 in human hematopoietic progenitors blocks erythroid differentiation induced by BMP4. Blood Cells Mol Dis. 2002;28:221–233. doi: 10.1006/bcmd.2002.0487. [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Stemmler MP. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J Biol Chem. 2003;278:3776–3785. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hooper AT, Butler JM, Nolan DJ, Kranz A, Iida K, Kobayashi M, Kopp HG, Shido K, Petit I, Yanger K, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide Y, Tsuchimoto D, Tominaga Y, Nakashima M, Watanabe T, Sakumi K, Ohno M, Nakabeppu Y. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 2004;104:4097–4103. doi: 10.1182/blood-2004-04-1476. [DOI] [PubMed] [Google Scholar]
- Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- Jeanpierre S, Nicolini FE, Kaniewski B, Dumontet C, Rimokh R, Puisieux A, Maguer-Satta V. BMP4 regulation of human megakaryocytic differentiation is involved in thrombopoietin signaling. Blood. 2008;112:3154–3163. doi: 10.1182/blood-2008-03-145326. [DOI] [PubMed] [Google Scholar]
- Koch U, Wilson A, Cobas M, Kemler R, Macdonald HR, Radtke F. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008;111:160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, Ten Dijke P, Heldin CH, Ericsson J, Moustakas A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol Cell Biol. 2003;23:4494–4510. doi: 10.1128/MCB.23.13.4494-4510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, Janes M, Piltz S, Hammett M, Carter J, et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- Larsson J, Karlsson S. The role of Smad signaling in hematopoiesis. Oncogene. 2005;24:5676–5692. doi: 10.1038/sj.onc.1208920. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chretien L, McKinney-Freeman S, Davidson AJ, Hammerschmidt M, Rentzsch F, Green JB, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- McReynolds LJ, Gupta S, Figueroa ME, Mullins MC, Evans T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood. 2007;110:3881–3890. doi: 10.1182/blood-2007-04-085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson KL, Hunter T, Janknecht R. Activation of Smad1-mediated transcription by p300/CBP. Biochim Biophys Acta. 1999;1489:354–364. doi: 10.1016/s0167-4781(99)00166-9. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F, Zhang J, Kramer C, Sebald W, Hammerschmidt M. Crossveinless 2 is an essential positive feedback regulator of Bmp signaling during zebrafish gastrulation. Development. 2006;133:801–811. doi: 10.1242/dev.02250. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlon TJ, Dell’Oso T, Surinya KH, May BK. Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis. Int J Biochem Cell Biol. 1999;31:1153–1167. doi: 10.1016/s1357-2725(99)00073-4. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Singbrant S, Karlsson G, Ehinger M, Olsson K, Jaako P, Miharada K, Stadtfeld M, Graf T, Karlsson S. Canonical BMP signaling is dispensable for hematopoietic stem cell function in both adult and fetal liver hematopoiesis, but essential to preserve colon architecture. Blood. 2010;115:4689–4698. doi: 10.1182/blood-2009-05-220988. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011 doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal FJ, Luis TC. Wnt signaling in hematopoiesis: crucial factors for self-renewal, proliferation, and cell fate decisions. J Cell Biochem. 2010;109:844–849. doi: 10.1002/jcb.22467. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Tran HT, Sekkali B, Van Imschoot G, Janssens S, Vleminckx K. Wnt/beta-catenin signaling is involved in the induction and maintenance of primitive hematopoiesis in the vertebrate embryo. Proc Natl Acad Sci U S A. 2010;107:16160–16165. doi: 10.1073/pnas.1007725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Winzeler A, Stern HM, Mayhall EA, Langenau DM, Kutok JL, Look AT, Zon LI. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood. 2004;104:1298–1305. doi: 10.1182/blood-2004-01-0100. [DOI] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61:800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- Tucker JA, Mintzer KA, Mullins MC. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc Natl Acad Sci U S A. 2010;107:15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. 2005;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book : a guide for the laboratory use of zebrafish (Danio rerio) Ed. 4. edn. Eugene, Or: M. Westerfield; 2000. pp. 1–35. [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.