Abstract
Pancreatic ductal adenocarcinomas (PDAC) are highly invasive and metastatic neoplasms commonly unresponsive to current drug therapy. Overwhelmingly, PDAC harbors early constitutive, oncogenic mutations in K-RasG12D that exist prior to invasion. Histologic and genetic analyses of human PDAC biopsies also exhibit increased expression of ERK1/2 and pro-invasive matrix metalloproteinases (MMPs); indicators of poor prognosis. However, the distinct molecular mechanisms necessary for K-Ras – ERK1/2 signaling and its influence on MMP-directed stromal invasion in primary human pancreatic ductal epithelial cells (PDECs) has yet to be elucidated in 3D. Expression of oncogenic K-RasG12D alone in genetically-defined PDECs reveals increased invadopodia and epithelial-to-mesenchymal transition markers, but only when cultured in a 3D model incorporating a basement membrane analog. Activation of extracellular signal-related kinase 2 (ERK2), but not ERK1, also occurs only in K-RasG12D mutated PDECs cultured in 3D and is a necessary intracellular signaling event for invasion based upon pharmacologic and shRNA inhibition. Increased active invasion of K-RasG12D PDECs through the basement membrane model is associated with a specific microarray gene expression signature and induction of MMP endopeptidases. Specifically, MMP-1 RNA, its secreted protein, and its proteolytic cleavage activity are amplified in K-RasG12D PDECs when assayed by RT q-PCR, ELISA, and fluorescence resonance energy transfer (FRET). Importantly, shRNA silencing of MMP-1 mimics ERK2 inhibition and disrupts active, vertical PDEC invasion. ERK2-isoform and MMP-1 targeting are shown to be viable strategies to attenuate invasion of K-RasG12D mutated human pancreatic cancer cells in a 3D tumor microenvironment.
Keywords: K-Ras, ERK2, Pancreatic Cancer, 3D Invasion, MMP-1
Introduction
Pancreatic cancer is fatal in 95% of patients within 6 months of diagnosis. This dismal prognosis is attributed to early tumor invasion, impalpable metastatic progression, and a lack of response to current therapeutic modalities (1,2). Pancreatic ductal adenocarcinoma (PDAC) accounts for the vast majority of exocrine pancreatic tumors and of these, 90% harbor early mutations in the K-Ras oncogene (3). The common single-nucleotide K-Ras mutation of G(G)X to G(A)X (glycine, G, to aspartate, D, respectively) at codon 12 (G12D) causes the membrane-associated GTPase K-Ras to remain locked in a constitutively GTP-bound state. In most epithelial cells, the activated K-RasG12D mutation constitutively signals through the MAPK cascade, increasing transcription of downstream genes. Many of these upregulated genes and their protein products enhance neoplastic growth, cytoskeletal arrangement, and metastasis (4,5). Considerable effort has been made to pharmacologically inhibit upstream effectors of this active K-Ras pathway, such as Raf and MEK, as well as Ras itself (6,7). Unfortunately, the complex signaling networks activated by mutant K-Ras differ between epithelial cell types. Thus, upstream effector targeting has limited downstream specificity resulting in toxicity and/or ineffectiveness (8,9). Considering that constitutive K-RasG12D is the most frequent mutation in pancreatic cancer and that invasion and metastasis of PDAC is early, insidious, and fatal, clinical advances will need clarification of the role of K-Ras in influencing invasion in a human model.
Genetically engineered mouse models of PDAC provide suitable systems capable of recapitulating the full progression of the human disease (10). These powerful models have furthered the understanding of K-Ras induced oncogenic effects but harbor deficits common to human translatability (10,11). Thus, groups have generated genetically engineered primary human pancreatic ductal epithelial cells (PDECs) through a series of defined, stepwise genetic alterations which cause transformation and result in anchorage-independent growth, augmented motility, hyperproliferation, and xenograftable tumors (12,13). These cell progression series incorporate human telomerase reverse transcriptase (hTERT), inactivation of Rb and p53 by the human papillomavirus E6 and E7 proteins, mutant K-RasG12D, and either SV40 small t or large T antigen. Although these in vitro human PDEC systems are capable of completely delineating the K-Ras controlled PI3K and Ral pathways, they have been unable to unequivocally show non-stimulated, constitutive K-Ras induced MAPK/ERK1/2 activation and transcriptional regulation in 2D (13,14). A need remains to determine the proteins, genes, and gene products under the influence of the mutated K-RasG12D – MAPK/ERK1/2 pathway in more physiologically relevant 3D basement membrane systems (15-17). The basement membrane and extracellular matrix (ECM) along with their inherent three-dimensionality greatly influence cell-cell contact, cell-matrix connections, cellular organization, intracellular signaling cascades, and gene expression in many ductal epithelial cells (15-20). Indeed, a highly inducible K-Ras – MAPK/ERK1/2 regulated gene expression pathway may also exist in pancreatic ductal cells and influence invasion when cultured in a 3D model (5).
Matrix metalloproteinases are a family of activated endopeptidases capable of enzymatically degrading ECM proteins that compose the tumor microenvironment. MMP signaling and remodeling of the local environment allows inherently motile transformed cells the ability to permissively invade into the surrounding tissue, eventually leading to metastasis (1,19-24). Human pancreatic cancer is consistently associated with a desmoplastic reaction incorporating increased levels of stromal and basement membrane ECM proteins (laminin, collagen IV), all of which are capable of degradation by MMPs (15,22-24). Specifically, the secreted collagenase, MMP-1, is found upregulated in pre-neoplastic pancreatitis and constitutively expressed within in vitro pancreatic cell lines (24,25). Catalytically active MMP-1 is capable of assisting in cell invasion either by cleavage of a G-protein-coupled receptor, PAR-1 (protease activated receptor-1), resulting in Rho cytoskeletal changes or by a mesenchymal type of invasion via connective tissue collagen and basement membrane degradation at a leading invadapodial edge, opening routes for metastasis through the ECM (21,22). Although signaling pathways controlling MMPs and their relative invasive importance in pancreatic cancer have been hypothesized, the exact molecular methods and genes necessary for K-RasG12D driven ECM invasion in human pancreatic cancer cells remains to be understood (26).
To identify the role of the K-RasG12D mutation in the morphology and invasion of pancreatic cancer, we characterized the mutation’s morphologic and invasive effects after addition into PDECs cultured within a 3D basement membrane model. Compared to untransformed PDECs, the K-RasG12D mutated cells exhibit increased invadapodia, epithelial-to-mesenchymal markers, and active, vertical basement membrane invasion in the synthetic stroma. To understand the mechanism of this invasiveness, we show that only mutated K-RasG12D PDECs signal specifically through ERK2, not ERK1, in 3D. The K-RasG12D – ERK2 signaling axis regulates invadapodia and active basement membrane invasion as well as a unique microarray gene expression signature distinct from non-mutated PDECs. Specifically, the constitutive activation of ERK2 by K-RasG12D is necessary for the upregulation of MMP-1 RNA, its secreted protein, and its proteolytic activity. Relevant to attenuating the early invasion and metastasis of PDAC, pharmaceutical inhibition or RNA silencing of either ERK2 or MMP-1 disrupts the vertical invasion of pancreatic cells into the 3D basement membrane.
Materials and Methods
Cell lines
Immortalized primary human pancreatic ductal epithelial cells (PDECs) were a generous gift from Dr. Michel Ouellette (University of Nebraska). All populations were maintained in conventional 2D cultures in T-75 flasks and a humidified incubator (95% air / 5% CO2) at 37°C. All PDECs were cultured in a pancreatic-specific growth medium: four parts low-glucose DMEM (Cellgro, Manassas, VA) to one part M3™ base culture medium (INCELL, San Antonio, TX) supplemented with 5% fetal bovine serum + 1% penicillin-streptomycin. Cells were passaged every 48 h by 0.05% trypsin detachment over 5 m.
Microarray Analysis
One microgram of isolated RNA was reverse transcribed to cDNA using the Qiagen FirstStrand Kit reaction assay on an Eppendorf Mastercycler Pro S (Hauppauge, NY). cDNA was aliquoted with Qiagen Mastermix and added to the wells of a SABiosciences Extracellular Matrix Microarray Plate (PAHS-0013, SABiosciences) at 25 μl/well. The cDNA plate was analyzed on an aluminum block Eppendorf ep Realplex II calibrated with an SABiosciences ‘A’ Plate using a 520-nm filter and a SYBR Green probe. The program contained a heat activation cycle followed by 40 cycles of PCR at a 35% ramp rate followed by one cycle of melting curve analysis to verify purity. Bioinformatic analysis by the Web-based RT2 Profiler PCR Array Data Analyzer (SABiosciences) was performed after logarithmic transformation of Ct amplification values against internal controls, while the threshold was consistently set. Data are expressed as ΔΔCt as well as fold-changes with ± 2-fold considered significant.
MMP activity assay
PDEC medium was collected after 24 h addition of either UO126, PD98059, LY294002, AEMT, or shRNA (ERK1, ERK2, MMP-1) lentiviral particles after 24 h in 3D culture. The activity of MMP-1 was assayed using a FRET based assay (AnaSpec, Fremont, CA, #71150) according to the manufacturer’s instructions: Briefly: a 5-FAM fluorescent molecule bound to a QXL™ 520 quencher by a cross-link capable of enzymatic cleavage specifically by MMP-1 was added to 25 μl of the collected PDEC medium. Upon separation from the quencher explicitly by MMP-1 enzymatic cleavage, the 5-FAM molecule fluoresces at 520 nm and the intensity correlates with MMP-1 activity. Pro-MMP-1 was activated immediately before the FRET MMP activity assay by incubating the collected PDEC medium with 5 μl of AMPA and kept on ice. Working solutions were prepared incorporating the MMP-1 substrate solution (a 5-FAM/QXL ™ 520 peptide) as well as the assay buffer. The enzymatic reaction was set up with two controls: 1.) a substrate control containing assay buffer and 2.) a positive control containing MMP-1 diluent. The fluorescence signal was measured kinetically every 5 m for 60 m at Ex/Em=490 nm /520 nm using a Synergy BioTek4 microplate reader.
Statistical analysis
All experiments were run in triplicate and repeated as stated. Where possible, the data are expressed as the average ± standard error of the mean (SEM). Significance was based upon p< 0.05 and 0.01 and clarified in each figure while error bars = SEM. Significance between two datasets was determined using a two-tailed Student’s t-test while differences between multiple experimental groups were determined by ANOVA with a Bonferroni post hoc test. Microarray datasets were initially filtered for genes with more than a two-fold change in Ct values. Internal microarray controls were determined across all samples for equal variance prior to comparison. Real-time quantitative polymerase chain reaction (RT q-PCR) determined significant increases in gene expression when sample RNA was used with specific Taqman™ probes by the Pffafl method.
Additional Materials and Methods are presented in the Supplementary Material.
Results
PDECs harboring a K-RasG12D mutation exhibit invasive morphology in 3D
Studies in various epithelial cells have shown that morphologic behavior depends on the dimensionality and signals provided by the surrounding ECM; thus a 3D model may more naturally mimic the endogenous milieu (15-17). In line with previous studies, all pancreatic ductal epithelial cells (PDECs) of the hTERT, hTERT plus E6 & E7 (E6/E7), and hTERT/E6/E7 plus mutated K-RasG12D (E6/E7/Ras) lineage exhibit similar fibroblast-like morphology in 2D, plastic culture, despite progressive inhibition of p53 and Rb or a constitutively active K-RasG12D mutation (12,13). Fluorescence microscopy shows similar F-actin cytoskeletal arrangements in all cell types maintained in 2D culture (Fig. 1A and Supplementary Fig. S1).
Figure 1. E6/E7/Ras PDECs exhibit invasive morphology in 3D culture.
(A) Schematic diagram representing the genetic steps taken to produce the clones of the progressively transformed PDEC cell series (as adapted from 13). Initial human telomerase reverse transcriptase (hTERT) next has the human papillomavirus E6 and E7 proteins retrovirally transfected followed by the K-RasG12D mutation; causing transformation. Retroviral transfection of K-RasG12D without the E6 and E7 proteins causes cellular senescence. (B) PDEC clones of the hTERT/E6/E7 plus mutant K-RasG12D lineage were cultured in 3D for 48 h and imaged by phase contrast or fluorescence microscopy. Phalloidin (green), DAPI (blue), and cortactin (red) antibodies were directed against F-actin, nuclei, and the invadapodia-specific and actin organizing protein. Single arrow = single cell invadopodia, double arrow = multicellular invadopodia. Scale bars = 50 microns. (C) Magnification of invadapodia of the E6/E7/Ras PDECs observed by phase contrast, DAPI (blue, nuclei), phalloidin (green, F-actin), and cortactin (red). Single arrow = single invadapodia, double arrow = multicellular invadapodia. Scale bars = 25 microns. (D) PDECs were cultured in 3D for 48 h and fluorescently labeled by antibodies directed against nuclei (DAPI, blue) and integrin alpha 6 (INTG6, red). Oncogenic K-RasG12D loses integrin alpha 6 attachment to Matrigel but increases the epithelial motility and invadapodial marker vimentin (DAPI, blue; Vimentin, red) Scale bars = 30 microns. (E) E6/E7 and E6/E7/Ras PDECs were cultured in 3D for 48 h prior to RIPA lysis and protein isolation. Protein samples were denatured, run on SDS-PAGE, and analyzed by Western Blot with an antibody directed against the intermediate filament, vimentin. Densitometric quantification of vimentin protein mimics increased vimentin expression by immunofluorescent staining. GAPDH was used as an internal normalization control. Scale bars = 20 microns. See also Supplementary Fig. S1 and Supplementary Movie S1A. All experiments: SEM and * = p< 0.05.
Interestingly, only culture in a 3D ECM basement membrane (Matrigel) model recapitulating pancreatic ductal architecture and elasticity (380 ± 63 Pa) reveals significant morphological differences between the three PDEC clones (Figs. 1B and 1C). hTERT and E6/E7 PDECs formed rounded, pseudo-organized multicellular aggregates, exhibiting a perimembranal web of F-actin. Moreover, the basolateral membrane of the hTERT or E6/7 PDEC clusters stained positive for integrin alpha 6, a laminin receptor (Fig. 1D). By contrast, E6/E7/Ras PDECs cultured in 3D displayed invasive morphology (stellate, invadopodia) characterized by similar cytoplasmic extensions found in metastatic epithelial cells (27-29). Individual invadopodia were composed of a single migrating cell extension (Fig. 1B and 1C, single arrow) or multicellular invadopodial aggregates extending from a central proliferative mass (Fig. 1B and 1C, double arrow and Supplementary Movie S1A). Phalloidin staining of the E6/E7/Ras PDECs exhibited an elongated F-actin distribution reminiscent of a mesenchymal phenotype. Further, while E6/E7 PDECs lacked nucleating F-actin markers of invadapodia, E6/E7/Ras PDECs where highly positive for the invadapodial marker cortactin (Figs. 1B and 1C). Magnification of individual invadapodia exhibit F-actin staining along the entire extension while parallel cortactin is found under the cell membrane as well as within developing invadapodial ‘buds’ (Fig 1C, see inset). Interestingly these cells lacked elaborate stress fibers and the surrounding basolateral membranes lacked integrin alpha 6 labeling (Figs. 1B and 1D). In addition to the presence of actin-rich invadopodial protrusions, vimentin, a mesenchymal intermediate filament necessary for invadapodial elongation and marker for a migratory epithelial cell phenotype was prominently upregulated in E6/E7/Ras PDECs over that of E6/E7 PDECs (Fig. 1C and D and 27-29). Taken together, these results demonstrate that a 3D ECM model composed of a basement membrane mimic is capable of revealing invasive morphologic and phenotypic differences between normal (E6/E7) and tumorigenic (E6/E7/Ras) pancreatic cancer cells.
Invasive phenotype of K-RasG12D pancreatic ductal epithelial cells is regulated by ERK2
We next attempted to identify downstream effector proteins involved in PDEC invasiveness and mediated by the K-RasG12D mutation. Neither the E6/E7 nor the E6/E7/Ras PDECs constitutively phosphorylated ERK1 or ERK2 proteins in 2D culture, despite the E6/E7/Ras PDECs possessing the constitutive, active K-RasG12D mutation (Supplementary Fig. S2). Addition of EGF transiently induced ERK1/2 phosphorylation, specifically only ERK2 in the E6/E7/Ras PDECs, which peaked after 30 m in 2D culture and returned to baseline levels within 1 h (Supplementary Fig. S3).
As only the E6/E7/Ras PDECs exhibited invasive morphology in 3D, we hypothesized that dimensionality might facilitate constitutive activation of downstream targets of mutated K-RasG12D. In contrast to 2D culture, ERK1 was phosphorylated in both cell types for at least 24 h in 3D culture even in the absence of EGF. Interestingly, constitutive phosphorylation of ERK2 occurred only in E6/E7Ras PDECs when cultured in 3D (Fig. 2A). A specific inhibitor of ERK’s upstream activator MEK (MAP2K), UO126, inhibited this constitutive ERK2 phosphorylation in 3D culture (Supplementary Fig. S4). As seen in Fig. 2B, ELK-1 (ets like gene-1) and RSK-1 (ribosomal S6 kinase-1), two downstream ERK substrates previously implicated in invasive gene transcription, were phosphorylated in E6/E7/Ras PDECs cultured in 3D (30-32). Phosphorylation of these two targets, but not of ERK2, was inhibited by 3-(2-aminoethyl)-5-((4-ethoxyphenyl) methylene)-2, 4-thiazolidinedione hydrochloride (AEMT), a small-molecule drug which preferentially antagonizes ERK2’s ability to phosphorylate these effectors (33). Importantly, E6/E7/Ras phosphorylation of RSK-1 could be reduced by 2.5-fold to the low-level, basal RSK-1 phosphorylation of E6/E7 PDECs by AEMT addition (Supplementary Fig. S5). Taken together, these experiments propose that 3D alone enables mutant K-RasG12D to specifically activate ERK2, not ERK1, signaling in PDECs.
Figure 2. E6/E7/Ras PDECs specifically signal through ERK2 in 3D.
(A) E6/E7 and E6/E7/Ras PDECs were cultured in 3D for 48 h prior to protein isolation after RIPA lysis. Western blot analysis determined the abundance of phosphospecific and total protein of ERK1 and ERK2 with specified antibodies. GAPDH was used as an internal normalization control. (B) 20 μM of the ERK2-specific inhibitor, AEMT, was added to both E6/E7 and E6/E7/Ras PDECs for 24 h after initial 3D culture. RIPA lysis and isolated protein samples of treated and untreated PDECs were probed for phosphospecific and total RSK-1, ELK-1, and ERK2. GAPDH was used as an internal normalization control. (C) To determine the influence of the PI3K pathway on PDEC growth and invasion, E6/E7/Ras PDECs cultured in 3D with or without epidermal growth factor (EGF) had 25 μM of the PI3K inhibitor LY294002 added for 24 h. Cell cultures were lysed by RIPA and had their protein isolated and probed with antibodies against total and phosphospecific Akt. EFG was added at a concentration of 10 μg/ml and GAPDH was used as an internal normalization control for densitometric quantification. There was no significant difference in the abundance of phosphorylated Akt before or after addition of EGF in either the E6/E7 or E6/E7/Ras PDECs. See also Supplementary Figs. S2, S3, and S4. All experiments: SEM and * = p< 0.05.
To assess if other activated K-Ras pathways are involved in 3D PDEC invasiveness, we examined phosphorylation of the PI3K-specific protein, Akt [T308], a known regulator of cell migration in PDECs (13). There was little difference in total or phosphorylated protein between E6/E7 and K-RasG12D mutated PDECs (Fig. 2C). The addition of EGF marginally upregulated E6/E7/Ras PDEC Akt phosphorylation, but was not significant, implying that K-RasG12D has greater influence on MEK-ERK2 signaling.
Morphologic characteristics were examined after treating the 3D PDECs with a panel of small-molecule inhibitors that are known to attenuate downstream Ras signaling pathways associated with transcription, motility, and cytoskeletal rearrangement (4). Strikingly, MEK inhibition with UO126 and PD98059 or ERK2-specific inhibition with AEMT drastically attenuated (in a dose and time-dependent manner) invadopodia and proliferation of the E6/E7/Ras PDECs in 3D (Figs. 3A and B). The PI3K inhibitor LY294002 did not significantly affect E6/E7/Ras invadopodial extensions in 3D, paralleling the lack of Akt phosphorylation observed in protein lysates. Importantly, at higher concentrations than used in this study, none of the small-molecule inhibitors affected viability in a variety of K-Ras mutated pancreatic cancer cells (Supplementary Fig. S6).
Figure 3. ERK2 regulates K-RasG12D mutated PDEC invasiveness.
(A) E6/E7/Ras PDECs were cultured in 3D for 24 h with the addition of DMSO (control), 20 μM of the PI3K pathway inhibitor (LY294002), 10 or 15 μM of MEK inhibitors (UO126 or PD98059, respectively), or 20 μM of the ERK2-specific inhibitor (AEMT). After applying the drugs for 24 h, each sample was observed by phase contrast microscopy. Scale bars = 20 microns. The number of invadopodia that developed after drug application were counted and normalized to E6/E7/Ras plus DMSO at 48 h. Each count is the combination of five areas of three independent experiments. Vertical bars represent SEM. (B) E6/E7/Ras PDECs were cultured in 3D with or without the ERK2-specific inhibitor AEMT or MEK inhibitor UO126 and imaged at 0 h and 48 h by phase contrast microscopy. Left scale bars = 50 microns, right scale bars = 100 microns. Inset = 10×. The number of invadopodia that developed after drug application were counted and normalized to E6/E7/Ras plus DMSO at 48 h. Each count is the combination of five areas of three independent experiments. Vertical bars represent SEM. (C) E6/E7/Ras PDECs were cultured upon growth factor reduced (GFR) Matrigel transwell invasion chambers for 48 h with DMSO (control), 20 μM AEMT, 10 μM UO126, 15 μM PD98059, or 20 μM LY294002. After, the bottom of each well’s membrane was fluorescently observed for the nuclei of invading cells. The number of invading E6/E7/Ras PDECs with DMSO addition was used as a normalization control for quantification. (D) MiaPaCa-2 cells, another human pancreatic cancer cell line harboring the K-Ras mutation, had 20 μM of the ERK2-specific inhibitor, AEMT, applied for 48 h on GFR Matrigel transwell invasion chambers and similarly analyzed for invasion. The number of invading MiaPaCa-2 cells with DMSO addition was used as a normalization control for quantification. See also Supplementary Fig. S6. All experiments: SEM and * = p< 0.05.
Because invadapodia were retracted following MEK or ERK2-specific inhibition, we tested if active invasion of E6/E7/Ras PDECs through a basement membrane was also inhibited. Indeed, K-Ras mutated PDECs and MiaPaCa-2 cells both exhibited decreased ECM invasion following MEK and ERK2-specific inhibition after 48 h (Figs. 3C and D). Overall, these results suggest that the invasive pancreatic cancer cell phenotype (invadapodia / active ECM invasion) is regulated by the constitutive activation of the K-RasG12D - ERK2 specific signaling axis in 3D.
ERK2 is necessary for an invasive PDEC phenotype
Recent publications implicate ERK2, not ERK1, as a potent inducer of epithelial-to-mesenchymal transition and suggest that its specific, active phosphorylation affects morphology, survival, and proliferation (30-35). To validate the necessity of ERK2 in the invasive phenotype of K-RasG12D mutated PDECs in 3D, we separately silenced endogenous ERK1 and ERK2 with two different shRNA lentiviral constructs (Fig. 4A). Silencing yielded at least a 70% decrease in both ERK isoforms in the E6/E7/Ras PDECs by either lentiviral construct. In agreement with the pharmacological inhibition studies, specific silencing of ERK2, but not ERK1, caused a significant reduction in the number of invadopodial protrusions observed in 3D (Fig. 4B). Additionally, silencing of ERK2, but not ERK1, substantially (by 80%) decreased the active invasion of the E6/E7/Ras PDECs through a basement membrane (Fig. 4C). Combined with the small-molecule inhibition results, the shRNA silencing data validates that ERK2, not ERK1, is the major isoform involved in the invasive regulation of K-RasG12D mutated pancreatic cells in 3D.
Figure 4. ERK2, not ERK1, silencing specifically decreases invasion of E6/E7/Ras PDECs in 3D.
(A) Two distinct shRNA expression constructs (A or B) for either ERK1 or ERK2 were introduced by lentiviral infection into E6/E7/Ras PDECs over 24 h. Scramble shRNA was introduced as a negative control while a GFP construct was added as a positive transfection control. At 24 h and 48 h post-infection, RIPA lysis of all cells was performed followed by protein isolation and Western Blot analysis. Lysates were probed for both total ERK1 and total ERK2 and quantified for protein abundance. GAPDH used as an internal normalization control. At least a 70% reduction in expression of ERK1 or ERK2 was observed in both the A and B silencing constructs. ** = p< 0.01 for ERK1 and ERK2 and vertical bars represent SEM. (B) Subpopulations of each E6/E7/Ras PDEC after silencing with sh-ERK1 or sh-ERK2 construct B were cultured in 3D for 48 h. Representative images of the populations after two independent experiments were observed by phase contrast microscopy. Scale bars = 20 microns. A minimum of 15 cell aggregates in five different images were counted and averaged for each condition after three independent experiments. (C) E6/E7/Ras PDECs were infected with lentiviral particles harboring scramble shRNA, sh-ERK1 B, or sh-ERK2 B for 24 h. PDECs were cultured inside transwell invasion plates covered in growth factor reduced (GFR) Matrigel for 48 h and nuclei were labeled with DAPI. Five individual areas for each shRNA construct of three separate experiments were counted for cells that invaded through the Matrigel to the opposite side of the membrane and compared with control. DMSO treated E6/E7/Ras PDECs were used as the normalized control of invasion. All experiments: SEM and * = p< 0.05 unless noted.
MMP-1 RNA expression is regulated by oncogenic K-RasG12D – MAPK/ERK2 signaling
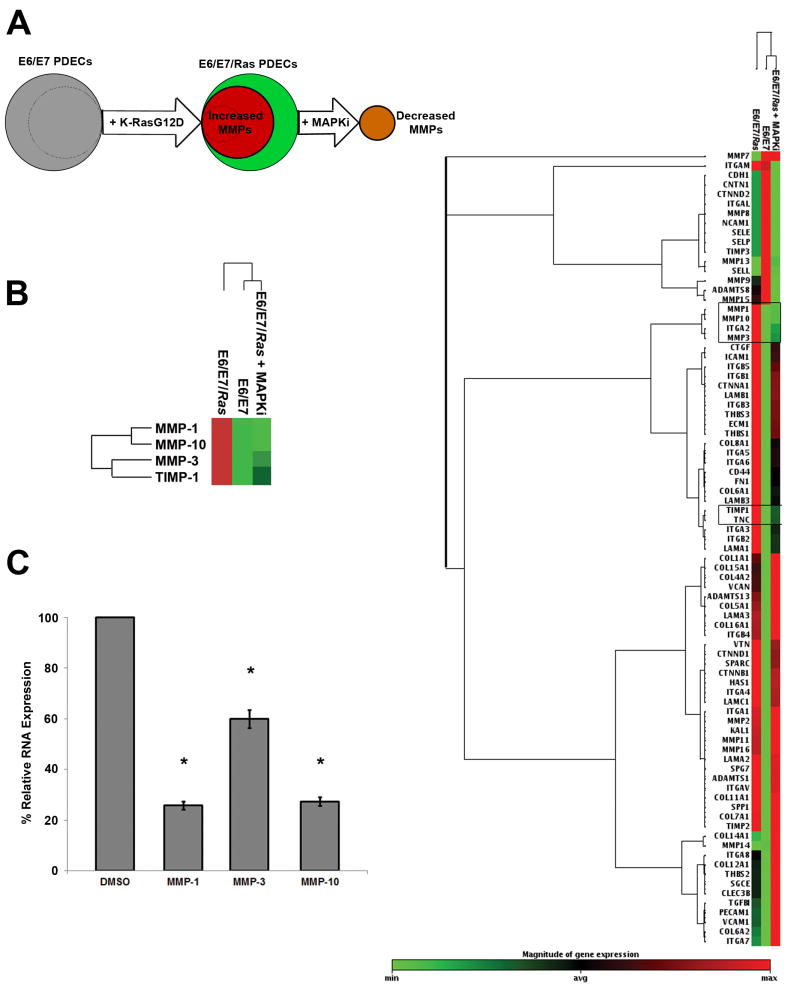
To identify genes stimulated by the activated K-Ras - MAPK/ERK2 signaling pathway, differential gene expression profiles of 3D cultured E6/E7 and E6/E7/Ras PDECs were generated by ECM-focused cDNA microarrays (Fig. 5A). While many genes were altered between the K-RasG12D transformed and untransformed E6/E7 PDECs, a specific subgroup of these genes was regulated exclusively by the MAPK pathway, as determined by microarray analysis after MAPK inhibition (MAPKi). Those genes which originally exhibited upregulation by K-RasG12D and then were subsequently downregulated following MAPKi were associated with a specific gene signature (Fig. 5B, left and right clustergram). The PDEC RNA microarray changes following MAPK pathway inhibition were validated by real time quantitative polymerase chain reaction (RT q-PCR, Fig. 5C). Many of the MAPK pathway regulated genes were of the MMP family: MMP-1, MMP-3, MMP-10, and tissue inhibitor of matrix metalloproteinase-1 (TIMP-1). Unbiased hierarchical analysis clustered E6/E7/Ras plus MAPKi with non-mutated E6/E7 PDECs and MMP-1, MMP-3, and MMP-10 within a closely linked node. Oncomine data mining of K-Ras mutated human pancreatic adenocarcinoma biopsy samples indicates similar upregulation of these MMPs after meta-analysis and Z-score calculation (Supplementary Fig. S7). Combined with previous data, these results support the notion that the K-Ras – MAPK/ERK2 pathway has a specific ECM invasion gene expression signature and upregulates specific MMP RNA in 3D.
Figure 5. E6/E7/Ras PDECs have a unique gene expression signature and regulate MMP genes.
(A) Schematic diagram representing the genetic techniques used in microarray analysis: K-RasG12D was retrovirally transfected into E6/E7 PDECs (as in Fig. 1A) and the RNA of both clones was isolated and purified. The subset of genes upregulated due to the mutation alone was then determined by microarray analysis. Genes that were significantly upregulated by the addition of the K-RasG12D transfection and subsequently reversed following MAPK pathway inhibition (MAPKi) were also determined after this RNA was isolated, purified, and run on the microarray. This permitted specific K-Ras – MAPK pathway regulated genes to be extracted for analysis. (B) Initially, the PDEC genes significantly upregulated by the K-RasG12D mutation (> 2-fold change, left) were extracted from the total microarray analysis of each experimental condition (right). Next, the genes subsequently downregulated after MAPKi were determined and consisted of MMP-1, MMP-3, MMP-10 and TIMP-1 (left and outlined in black framed boxes in total microarray). (C) RNA from E6/E7/Ras PDECs was isolated after 48h culture in 3D conditions, purified, and mixed with MMP-1, MMP-3, or MMP-10 Taqman gene expression probes. Real-time quantitative polymerase chain reaction (RT q-PCR) was able to validate the microarray results by also determining the increased expression of these RNA transcripts in E6/E7/Ras PDECs. GAPDH was used as an internal ‘housekeeping’ control and all Ct values were normalized to its expression. Analysis of significant gene expression increases was determined by the Pffafl method and ANOVA with a Bonferroni post hoc test. All experiments: SEM and * = p< 0.05.
MMP-1 protein and invasive activity are specifically regulated by the K-Ras - ERK2 pathway
In order to ascertain which of the transcriptionally upregulated MMPs contribute to the invasiveness of E6/E7/Ras PDECs in 3D, the effects of mutated K-RasG12D on MMP-1, MMP-3, and MMP-10 zymogen release was studied. Incorporating ELISA, medium from E6/E7 and E6/E7/Ras PDECs was analyzed for the concentration of specific MMPs after 48 h in 3D culture. Although RNA for each of the three MMPs had increased (Fig. 5) and all are extracellularly secreted, only MMP-1 protein was significantly detectable by ELISA (Fig. 6A). The abundance of MMP-1 protein was significantly higher (3-fold) in the E6/E7/Ras cells over the non-mutated PDECs (Fig. 6B and Supplementary Fig. S8). Addition of EGF lead to ERK2 phosphorylation and increased MMP-1 secretion in E6/E7/Ras but not in non-mutated E6/E7 PDECs, mimicking both 2D plus EGF and 3D-only culture conditions (Fig. 6B and Supplementary Fig. S3). Conversely, inhibition of MEK or ERK2 signaling with UO126 or AEMT, respectively, significantly decreased MMP-1 secretion in K-RasG12D mutated PDECs (Fig. 6B). Lastly, shRNA silencing of ERK2, but not ERK1, also significantly decreased MMP-1 zymogen secretion from 3D cultured PDECs (Fig. 6C).
Figure 6. Secreted and active MMP-1 protein is regulated by the K-RasG12D - ERK2 pathway.
(A) Medium was collected from E6/E7/Ras PDECs after 48 h in 3D culture to determine the total protein concentration of MMP-1, MMP-3, and MMP-10 secreted into the extracellular environment by enzyme linked immunosorbent assay (ELISA). The total MMP protein concentration was extrapolated from the fluorescent measurements of known human MMP-1 serial dilutions set to a standard curve. ** = p< 0.01. (B) Total MMP-1 protein secreted into the extracellular environment by E6/E7 and E6/E7/Ras PDECs in 3D was quantified following treatment with EGF, UO126, and AEMT over 48 h by ELISA. The EGF concentration = 10 ng/ml, UO126 = 15 μM, and AEMT = 20 μM. DMSO was used as a vehicle control. (C) Total secreted MMP-1 protein was determined by ELISA following RNA silencing by sh-ERK1 B or sh-ERK2 B in 3D cultured E6/E7/Ras PDECs 48 h after knockdown. Silencing ERK2 reduced the total secreted abundance of MMP-1 by approximately 50% compared with only an 8% reduction upon ERK1 silencing. (D) Medium sampled from 3D cultures of E6/E7 or E6/E7/Ras PDECs after a 48 h addition of the MEK inhibitor, UO126, ERK2 specific inhibitor, AEMT, or sh-MMP-1 lentiviral particles was collected for analysis. A 5-FAM-quencher FRET peptide was added to each collected sample, pro-MMP-1 was activated by AMPA, and the enzymatic ability of MMP-1 to cleave the 5-FAM molecule from the quencher was read over 60 m as per the Materials and Methods. MMP-1 activity was analyzed kinetically at 5 m time points by fluorescence emitted from the freed 5-FAM molecule. Vertical bars indicate SEM and significant differences in MMP-1 activity between E6/E7 (— -), E6/E7/Ras (——), E6/E7/Ras plus AEMT (- - -), and E6/E7/Ras plus UO126 (— —), and E6/E7/Ras + sh-MMP-1 (- - —) occurred at 35 m post-activation and extend to 60 m. See also Fig. S8. All experiments: SEM and * = p< 0.05 unless noted.
Because ELISA quantifies total secreted proteases (inactive and active) and considering that only activated MMPs are capable of proteolytic PDAC ECM degradation, zymography and FRET were employed to quantify the enzymatic activity rate of MMP-1 secreted from 3D PDECs (22). In line with MMP-1 ELISA results, kinetic analysis verified MMP-1 proteolysis was augmented in E6/E7/Ras medium compared to E6/E7 PDECs and was readily reversed following MEK or ERK2 inhibition (Fig. 6D). E6/E7/Ras PDECs exhibited an average MMP-1 enzymatic activity rate of 33.4 ± 0.25 Arbitrary Units per minute (A.U./minute) while the E6/E7 PDEC activity rate was 5-fold lower (6.0 ± 0.3 A.U./minute). Treatment of 3D E6/E7/Ras PDECs with the MEK or ERK2-specific inhibitors (UO126 or AEMT) significantly lowered their protease activity rate to that of E6/E7 PDECs over 1 h while silencing of MMP-1 demonstrated a proteolytic rate that was inherently indistinguishable from the E6/E7 PDECs (Fig. 6D). Thus, the K-RasG12D mutation increases MMP-1 transcription, protein secretion, enzymatic activity, and invasion of 3D cultured PDECs by a MEK-ERK2 signaling axis.
MMP-1 is necessary for K-RasG12D mutated PDEC invasion
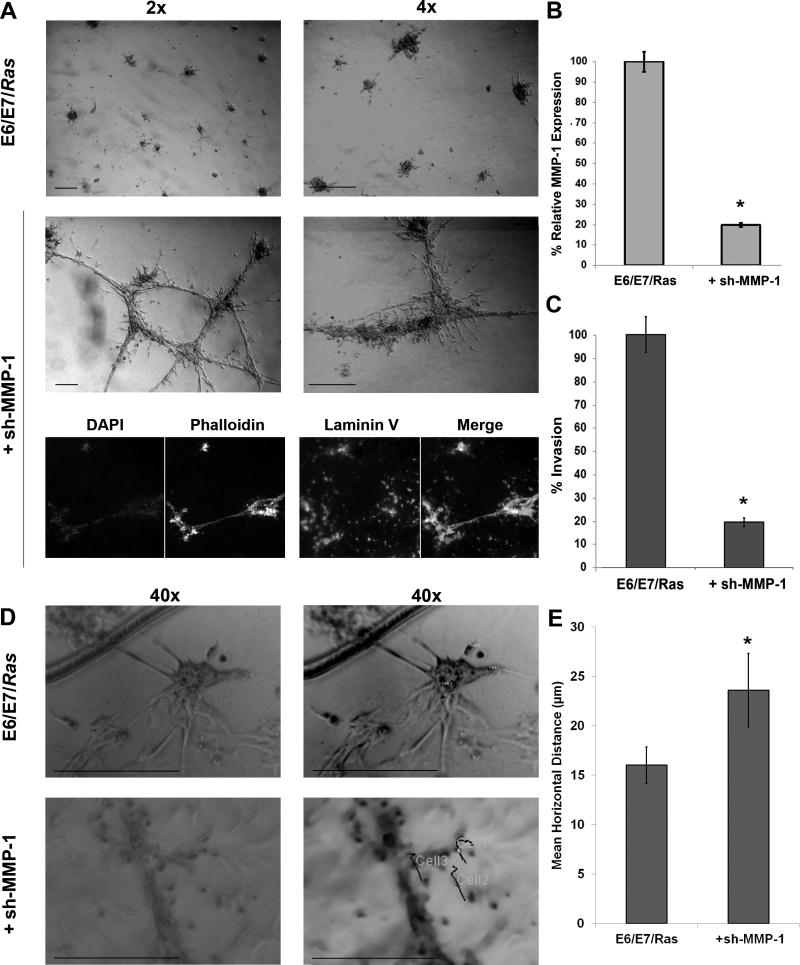
To determine the K-Ras regulated MMP-1 influence on basement membrane invasion, E6/E7/Ras PDEC MMP-1 RNA was silenced by shRNA lentivirus. As seen in Fig. 7A, the E6/E7/Ras PDECs with the addition of five pooled sh-MMP-1 constructs exhibited increased horizontal spreading across the ECM when compared to control over 30 m. Fluorescent phalloidin imaging shows multicellular extensions along the length of trabecular structures terminating in laminin-V rich areas, common to migratory pancreatic cells (36). RT q-PCR analysis established that MMP-1 was approximately 80% silenced compared to control (Fig. 7B). Importantly, MMP-1 silencing in E6/E7/Ras PDECs by shRNA reduced its protease activity to that of E6/E7 PDECs over 1 h (Fig. 6D). Further, basement membrane invasion assays determined that the sh-MMP-1 E6/E7/Ras PDECs had an approximately 80% reduction in vertical invasion through the Matrigel basement membrane, as would be expected with its decreased MMP-1 expression and protease activity (Fig. 7C). In agreement, live, individual cell motion tracking of E6/E7/Ras PDECs with and without sh-MMP-1 lentiviral particle infection determined a significant increase in the total horizontal distance traveled by E6/E7/Ras PDECs when MMP-1 was silenced (Figures 7D and 7E and Supplementary Movies S2A and S3A). Therefore, specific inhibition of MMP-1 impedes active vertical invasion into the ECM and subsequently facilitates the horizontal spreading of inherently migratory K-RasG12D PDECs upon the basement membrane.
Figure 7. MMP-1 silencing attenuates vertical E6/E7/Ras ECM invasion.
(A) E6/E7/Ras PDECs were subjected to lentiviral particles carrying five pooled sh-MMP-1 expression constructs for 24 h before being added to 3D culture for another 24 h. Cells were fixed with 5% formalin prior to phase and fluorescent imaging. Antibodies towards phalloidin (green), DAPI (blue), and human laminin V (red) were applied with appropriate fluorescent secondary antibodies to visualized F-actin polymerization, nuclei and cellular deposition of human laminin upon the Matrigel using a Nikon microscope. (B) The efficiency of MMP-1 silencing following lentiviral infection of an shRNA construct was determined by real-time quantitative polymerase chain reaction (RT q-PCR) and normalized to GAPDH. (C) E6/E7/Ras PDECs with and without introduction of sh-MMP-1 shRNA were cultured on GFR Matrigel invasion chambers for 48 h, DAPI labeled, and counted. Five individual areas of three independent experiments were counted and the invasion percentage was determined by dividing the number of invading PDECs after silencing by those invading in the control. (D) E6/E7/Ras PDECs with and without sh-MMP-1 lentiviral particle infection were cultured within 3D GFR Matrigel for 24 h and live-imaged over 15 h at 37°C and 5.5% CO2 as per the Materials and Methods. Original image stacks (left) were normalized for pixel and magnification and then processed by the Circadian Gene Expression (CGE) ImageJ plug-in (right) to determine individual cells. The CGE recorder was then utilized to track the movement of three cells in five different areas of repeated experiments over 30 min (Cell1, Cell2, Cell3) (E) After cell tracking, each traces’ measurements were sent to a spreadsheet where total horizontal distance traveled was determined by summation of each frame’s hypotenuse; determined from the cell’s X and Y coordinate in the preceding frame. See also Supplementary Materials and Methods and Supplementary Movies S2A and S3A. All experiments: scale bars = 100 microns; SEM and * = p< 0.05.
Overall, these data indicate that the specific addition of a K-RasG12D mutation to human PDECs increases invasive characteristics via activation of the MAPK-ERK2 pathway. In turn, the constitutive activation of the ERK2-specific signaling axis regulates a unique gene expression signature in PDECs and increases MMP RNA transcripts. Distinct increases in MMP-1 gene expression correlates with amplified secretion of enzymatically active MMP-1 protein which is necessary for invasion into the basement membrane by human pancreatic cancer cells.
Discussion
Three main messages emerge from this study: 1.) 3D culture of human PDECs reveals a distinct invasive phenotype regulated by mutated K-RasG12D alone, which is not detected in conventional 2D culture, 2.) Invasion is specifically mediated by an active ERK2 signaling pathway and is detected only in 3D and only in K-Ras mutated pancreatic cancer cells, and, 3.) Constitutive activation of MAPK - ERK2 increases transcription, protein abundance, and activity of MMP-1 which is necessary for pancreatic cancer cell invasion in 3D. Taken together, this set of data is important as the significance of the K-Ras - MAPK - ERK2 - MMP pathway in mutated K-RasG12D pancreatic cancer cells has not been previously demonstrated. Prior 2D studies found that constitutively active K-Ras phosphorylated MEK but not ERK unless growth factors were added to pancreatic cells (13,14,37). The present study underscores the idea that proper assessment of signaling pathways, specifically through ERK2, in cultured cells requires emulation of the appropriate 3D physiologic environment of the pancreas, namely the addition of basement membrane components (5,38,39).
E6/E7/Ras PDECs harboring the constitutively active K-RasG12D mutation exhibit invadopodia and increased cell numbers per aggregate similar to transformed prostate and mammary epithelial cells in 3D (Fig. 1, Supplementary Movie S1A, 39-41). Importantly, preliminary data suggests that this morphologic differentiation is only possible in Matrigel and not in collagen type I. Insertion of a K-RasG12D mutation alone in hTERT PDECs results in senescence (13). Therefore, immortalized PDECs require p53 ubiquitin-mediated degradation and p16INK4a/Rb inactivation (by E6 and E7 genes, respectively) in addition to the K-RasG12D mutation for this transformation. Non-mutated E6/E7 PDECs exhibit a well-developed system of stress fibers and peripheral integrin alpha 6 expression in 3D, while the K-RasG12D mutation causes aggregate disorganization and decreased expression of this laminin binding integrin; common characteristics of invasive pancreatic cells (42). Correlatively, the invadapodia-specific markers cortactin and vimentin exhibited increased fluorescent expression in E6/E7/Ras PDECs, underlining true invadapodial development. In addition to increased cortactin and vimentin labeling, intercellular vimentin protein also increased in the E6/E7/Ras PDECs, all of which are seen in EMT and pancreatic cell invasion (27-29, 41,43). Evaluation of the ECM microarrays of the different PDEC clones indicates upregulation of several other EMT associated genes besides these in E6/E7/Ras PDECs (Supplementary Fig. S9). CD44, a hyaluronan receptor associated with cancer stem cells and malignancy, was upregulated along with the EMT markers fibronectin, integrin β2, tenascin-c, and vitronectin (43,44). As addition of a K-RasG12D mutation is the sole genetic distinction between E6/E7 and E6/E7/Ras PDECs and mutated cells have significant 3D invasive characteristics, we conclude that constitutively active K-Ras is the key factor regulating these invasion-inducing downstream genes.
Ras regulates gene expression pathways controlling motility, invasion, and proliferation via downstream effector pathways. Considering the influence of K-Ras on the MAPK pathway, phosphorylated ERK1/2 is upregulated in surgically resected K-Ras mutated pancreatic cancer specimens (43,45). In 2D culture, EGF induces ERK1 phosphorylation in PDECs regardless of their K-RasG12D mutational status. Interestingly, EGF causes phosphorylation of ERK2 only in K-RasG12D mutated PDECs (Supplementary Fig. S3). Remarkably, 3D culture (in the absence of exogenous EGF) is the only factor necessary to mimic these results in E6/E7/Ras PDECs (Fig. 2). Specific inhibition of ERK2 by AEMT or shRNA (Figs. 3, 4 and Supplementary Fig. S5) decreases K-RasG12D downstream signaling as well as E6/E7/Ras and MiaPaCa-2 invasiveness suggesting that 3D plus the iterative K-RasG12D mutation is sufficient to induce ERK2 phosphorylation/activation and invasion.
Various Ras mutants (H-RasG12V / K-RasG12V / K-RasG13D) constitutively activate MAPK pathway signaling and subsequently increase genes that mediate epithelial cell invasion (30,32). Specific regulation of RSK/FRA-1 (fos related antigen-1) interactions by the D-domain of ERK2 has been shown in kidney cells and breast and colon cancer cells (32). Additionally, ERK2 is recognized as an isoform-specific regulator of EMT through DEF-motif docking sites in breast cancer cells (30). DEF-domain EMT is characterized by invasive morphologic changes, decreased E-cadherin expression, and increased expression of vimentin, N-cadherin, and fibronectin - similarly determined in the pancreatic cancer cells of this study (Fig. 1C and D, Supplementary Fig. S9, and 30,43). Both the ERK2 D-domain and DEF-domain substrates seem significant to K-RasG12D mutated PDEC signaling in 3D, since neither ELK-1 (DEF- and D-domains) nor RSK-1 (DEF-domain) were significantly phosphorylated in E6/E7 PDECs or following ERK2 inhibition in E6/E7/Ras PDECs by AEMT (an inhibitor of both motifs, Fig. 2B and Supplementary Fig. S5). As ERK1 knockout mice survive past birth while ERK2 knockout mice die en utero, it follows that ERK2 may direct unique signaling traits over ERK1 (35).
Specific ERK2 signaling in 3D may derive from focal adhesion stabilization of a unique intercellular scaffold protein. The scaffolding proteins FAK, KSR (kinase suppressor of Ras), Sef (similar expression to FGF genes), and IQGAP1 (IQ motif containing GTPase activating protein 1) increase MAPK signaling efficiency by facilitating proper gathering and orientation of effectors (46,47). A preferential ERK2-binding scaffold is plausible considering the discovery of the preferential MEK1 - ERK1 binding scaffold, MEK Partner 1 (MP1, 48). Differences seen in F-actin stress fiber formation and cortactin localization within 3D cultured PDECs support this hypothesis (Fig. 1A and 46-48).
Microarray analysis identified distinct ECM-related genes that are increased in K-RasG12D PDECs over E6/E7 when cultured in 3D (Fig. 5). When E6/E7/Ras PDECs are then inhibited by MAPKi, we observed decreased expression of genes that are specifically controlled by this pathway: MMP-1, MMP-3, MMP-10, and TIMP-1. Unbiased clustering analysis more similarly separates the E6/E7 and E6/E7/Ras plus MAPKi PDEC gene expression profiles apart from E6/E7/Ras, signifying their similar genetic regulation (Fig. 5B). Meta-analysis of data mined from human pancreatic adenocarcinoma biopsy samples agrees with the upregulation of these MMPs within K-Ras mutated tumors, further pinpointing their influence (Supplementary Fig. S7). Interestingly, MMP-1, MMP-3, and MMP-10 all reside on chromosome 11 and have similar promoter elements enriched in activator protein 1 (AP-1) and polyoma enhancing activator 3 (PEA-3) binding sites approximately -180 bp upstream from the transcription start site (49). Coincidently, MMP-1 expression is regulated by MAPK-related transcription factors (c-fos and c-jun) at these sites in gastric cancers (50). MMP-7 and MMP-14 (MT-MMP-1) have been implicated in the invasion of pancreatic cancer cells and PDAC (51,52). Interestingly, the expression of these MMPs is higher in E6/E7 and in E6/E7/Ras plus MAPKi PDECs at 48 h, implying later upregulation in K-RasG12D PDECs.
Broad-spectrum MMP inhibitors were clinically withdrawn due to unforeseen complexities in the influence of MMPs on tumor progression and inhibition (53). It is necessary to understand the tissue specificity and the proper balance of MMPs in various pathologies as many MMPs are effective tumor suppressors and their inhibition can expedite neoplastic development. In this study, increasing levels of MMP-1 RNA correlated to increases in secreted MMP-1 protein and activity but only in 3D E6/E7/Ras PDECs and not when MMP-1 RNA was silenced (Fig. 6 and Supplementary Fig. S8). Although MMP-3 and MMP-10 transcripts were also increased in the medium of E6/E7/Ras PDECs, their protein secretion was not measurable by ELISA signifying post-transcriptional attenuation or extracellular neutralization, possibly by TIMPs. Kinetic analysis of MMP-1 protease activity using a FRET assay demonstrated that MMP-1 secretion/activity could be reduced to non-K-RasG12D mutated levels with pharmacologic and shRNA inhibition of ERK2, but not ERK1. Further, shRNA silencing of MMP-1 in E6/E7/Ras PDECs by 80% reduced its protease activity to that of the E6/E7 PDECs without the K-RasG12D mutation, thereby underlining this mutation’s influence in increasing the abundance of mRNA, protein, and enzymatic activity of MMP-1 (Figs. 5 and 6). Silencing MMP-1 also disrupted the ability of the cells to vertically invade into the basement membrane and instead significantly redirected their intrinsic motility across the Matrigel surface horizontally (Fig. 7, Supplementary Movies S2A and S2B). Thus, the upregulation of MMP-1 by K-RasG12D as well as its downregulation after ERK2 and shRNA inhibition directly links the mutated K-RasG12D - ERK2 signaling pathway to the MMP-1 mediated invasiveness of PDECs in 3D. MMP-1 expression may be increased due to interactions with the laminin, proteoglycans, and collagen IV present in the human basement membrane, aiding in their degradation as seen in Matrigel models (54, 55, 56, 57). Further examination of collagen IV proteolysis by MMP-1 seems appropriate. Although silencing MMP-1 in E6/E7/Ras PDECs reduces their invasion into the ECM and unveils their inherent horizontal motility rate compared to the same clone without sh-MMP-1, we cannot definitively rule out the possibility that MMP-1 may be cleaving PAR-1, assisting in invasion (21). The positive F-actin, vimentin, and cortactin labeling as well as elongated morphology, invadapodia extension, and increased protease activity of E6/E7/Ras PDECs implies the necessity of the Arp2/3 complex in directing mesenchymal, invadapodial invasion and runs contrary to an amoeboid migration model of PDECs through Matrigel (Figure 1 and Supplementary Movies S2A, S2B and 58). Overall, the results of this study substantiate that the K-RasG12D regulated ERK2 pathway and subsequent MMP-1 induction are specific therapeutic targets potentially capable of decreasing the stromal invasiveness of pancreatic adenocarcinoma.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Drs. Michel Ouellette (University of Nebraska), Jane Clifford, Gianluca Gallo, Boris Polyak, Shimon Lecht (Drexel Medicine), Maureen Murphy (Fox Chase Cancer Institute), John Blenis (Harvard Medical School), and their labs, the Penn Pancreatic Cancer Group, and the entire Drexel iCTERM Center for their assistance. This work was supported in part by the NIDDK: F30 DK088402-01 to G.P.B., the NCI: R01 CA155413-01 to M.J.R., the NIH: R01 DK060694 to A.K.R., and the NTI to P.I.L.
References
- 1.Keleg S, Büchler P, Ludwig R, Büchler MW, Friess H. Invasion and metastasis in pancreatic cancer. Mol Cancer. 2003;2:14. doi: 10.1186/1476-4598-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi AK. The molecular pathogenesis of pancreatic cancer: clarifying a complex circuitry. Genes Dev. 2006:3049–53. doi: 10.1101/gad.1501106. [DOI] [PubMed] [Google Scholar]
- 4.Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, et al. Genetic analysis of Ras Signaling pathways in cell proliferation, migration, and survival. EMBO. 2010:1091–1104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowy AM, Leach SD, Philip P. Pancreatic Cancer. New York: Spring Science + Business Media, LLC; 2008. [Google Scholar]
- 6.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 7.Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochim Biophys Acta. 2005:127–44. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao YM, et al. PI3K Pathway Activation Mediates Resistance to MEK Inhibitors in KRAS Mutant Cancers. Cancer Res. 2009:4286. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 9.Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. PNAS. 2010:1–6. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007:645–58. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 11.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commu. 2003:1038–44. doi: 10.1016/s0006-291x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ, et al. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and Phosphatiylinositol 3-Kinase Signaling. Cancer Res. 2007:2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 14.Yip-Schneider MT, Lin A, Barnard D, Sweeney CJ, Marshall MS. Lack of elevated MAP kinase (Erk) activity in pancreatic carcinomas despite oncogenic K-ras expression. Int J Oncol. 1999:271–279. doi: 10.3892/ijo.15.2.271. [DOI] [PubMed] [Google Scholar]
- 15.Nelson CM, Bissell MJ. Modeling dynamic reciprocity: Engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin Cancer Biol. 2005:342–52. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botta GP, Manley P, Miller S, Lelkes PI. Real-time assessment of three-dimensional cell aggregation in rotating wall vessel bioreactors in vitro. Nat Protoc. 2006:2116–27. doi: 10.1038/nprot.2006.311. [DOI] [PubMed] [Google Scholar]
- 17.Yamada KM, Cukierman E. Modleling Tissue Morphogenesis and Cancer in 3D. Cell. 2007:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Mahadevan D, von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2007:1186–97. doi: 10.1158/1535-7163.MCT-06-0686. [DOI] [PubMed] [Google Scholar]
- 19.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Brien LE, Tang K, Kats ES, Schutz-Geschwender A, Lipschutz JH, Mostov KE. ERK and MMPs Sequentially Regulate Distinct Stages of Epithelial Tubule Development. Dev Cell. 2004:21–32. doi: 10.1016/j.devcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Yang E, Boire A, Agarwal A, et al. Blockade of PAR1 signaling with cell-penetrating pepducins inhibits Akt survival pathways in breast cancer cells and suppresses tumor survival and metastasis. Cancer research. 2009;69(15):6223–31. doi: 10.1158/0008-5472.CAN-09-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimi S, Shono T, Ono M, Kuwano M, Tanaka M, Lopezotin C, Kono A. Expression of matrix metalloproteinases 1 and 2 genes in a possible association with metastatic abilities of human pancreatic cancer cells. Int J Oncol. 1997:623–8. doi: 10.3892/ijo.10.3.623. [DOI] [PubMed] [Google Scholar]
- 23.Chu GC, Kimmelman AC, Hezel AF, DePinho RA. Stromal biology of pancreatic cancer. J Cell Biochem. 2007:887–907. doi: 10.1002/jcb.21209. [DOI] [PubMed] [Google Scholar]
- 24.Nakae H, Endo S, Inoue Y, Fujino Y, Wakabayashi G, Inada K, et al. Matrix metallo-proteinase-1 and cytokines in patients with acute pancreatitis. Pancreas. 2003:134–8. doi: 10.1097/00006676-200303000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, et al. Highly Expressed Genes in Pancreatic Ductal Adenocarcinomas: A Comprehensive Characterization and Comparison of the Transcription Profiles Obtained from Three Major Technologies. Cancer Res. 2003:8614–8622. [PubMed] [Google Scholar]
- 26.Endo H, Watanabe T, Sugioka Y, Niioka M, Inagaki Y, Okazaki I. Activation of two distinct MAPK pathways governs constitutive expression of matrix metalloproteinase-1 in human pancreatic cancer cell lines. Int J Oncol. 2009:1237–45. doi: 10.3892/ijo_00000440. [DOI] [PubMed] [Google Scholar]
- 27.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial Actin Dynamics Control Epithelial-Mesenchymal Transition in Metastatic Cancer Cells. Cancer Res. 2010:3780–90. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 28.Gilles C, Polette M, Zahm JM, Tournier JM, Volders L, Foidart JM, et al. Vimentin contributes to human mammary epithelial cell migration. J Cell Sci. 1999:4615–25. doi: 10.1242/jcs.112.24.4615. [DOI] [PubMed] [Google Scholar]
- 29.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadapodia. J Cell Biol. 2010:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin S, Dimitri C, Yoon SO, Dowdle W, Blenis J. ERK2 but Not ERK1 Induced Epithelial-to-Mesenchymal Transformation via DEF Motif-Dependent Signaling Events. Mol Cell. 2010:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burkhard KA, Chen F, Shaprio P. Quantitative analysis of ERK2 interactions with substrate proteins, roles for kinase docking domains and activity in determining binding affinity. J Biol Chem. 2011:2477–85. doi: 10.1074/jbc.M110.177899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, Cohen MS, et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/ invasive gene program and phenotype in epithelial cells. Mol Cell. 2009:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock CN, Macias A, Lee EK, Yu SY, Mackerell AD, Jr, Shapiro P. Identification of Novel Extracellular Signal-Regulated Kinase Docking Domain Inhibitors. J Med Chem. 2005:4586–4595. doi: 10.1021/jm0501174. [DOI] [PubMed] [Google Scholar]
- 34.Vantaggiato C, Formentini I, Bondanza A, Bonini C, Naldini L, Brambilla R. ERK1 and ERK2 mitogen-activated protein kinases affect Ras-dependent cell signaling differentially. J Biol. 2006:1–15. doi: 10.1186/jbiol38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefloch R, Pouyssegur J, Lenormand P. Single and Combined Silencing of ERK1 and ERK2 Reveals Their Positive Contribution to Growth Signaling Depending on Their Expression Levels. Mol Cell Biol. 2008:511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jesnowski R, Fürst D, Ringel J, Chen Y, Schrödel A, Kleef J, et al. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by Matrigel and N-acetylcysteine. Lab Invest. 2005:1276–91. doi: 10.1038/labinvest.3700329. [DOI] [PubMed] [Google Scholar]
- 37.Veit C, Genze F, Menke A, Hoeffert S, Gress TM, Gierschik P. Activation of phosphatidylinositol 3-kinase and extracellular signal-regulated kinase is required for glial cell line-derived neurotrophic factor-induced migration and invasion of pancreatic carcinoma cells. Cancer Res. 2004:5291–300. doi: 10.1158/0008-5472.CAN-04-1112. [DOI] [PubMed] [Google Scholar]
- 38.Nelson CM, Bissell MJ. Of Extracellular Matrix, Scaffolds, and Signaling; Tissue Architecture Regulates Development, Homeostasis, and Cancer. Annu Rev Cell Dev Biol. 2006:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webber MM, Bello D, Kleinman HK, Hoffman MP. Acinar differentiation by non-malignant immortalized human prostatic epithelial cells and its loss by malignant cells. Carcinogenesis. 1997:1225–31. doi: 10.1093/carcin/18.6.1225. [DOI] [PubMed] [Google Scholar]
- 41.Tolde O, Rosel D, Vesely P, Folk P, Brabek J. The structure of invadopodia in a complex 3D environment. J Cell Biol. 2010:674–680. doi: 10.1016/j.ejcb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Walsh N, Clynes M, Crown J, O’Donovan N. Alterations in integrin expression modulates invasion of pancreatic cancer cells. J Exp Clin Cancer Res. 2009:140–152. doi: 10.1186/1756-9966-28-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javle MM, Gibbs JF, Iwata KK, Pak Y, Rutledge P, Yu J, et al. Epithelial-Mesenchymal Transition (EMT) and Activated Extracellular Signal-regulated Kinase (p-ERK) in Surgically Resected Pancreatic Cancer. Ann Surg Oncol. 2007:3527–33. doi: 10.1245/s10434-007-9540-3. [DOI] [PubMed] [Google Scholar]
- 44.Blick T, Hugo H, Widodo E, Waltham M, Pinto C, Mani SA, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia. 2010:235–252. doi: 10.1007/s10911-010-9175-z. [DOI] [PubMed] [Google Scholar]
- 45.Hoshino R, Chatani Y, Yamori T, Tsuruo T, Oka H, Yoshida O, et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999:813–22. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 46.Dhanasekaran DN, Kashef K, Lee CM, Xu H, Reddy EP. Scaffold Proteins of MAP-kinase Modules. Oncogene. 2007:3185–3203. doi: 10.1038/sj.onc.1210411. [DOI] [PubMed] [Google Scholar]
- 47.Sawai H, Okada Y, Funahashi H, Matsuo Y, Takahashi H, Takeyama H, et al. Activation of focal adhesion kinase enhances the adhesion and invasion of pancreatic cancer cells via extracellular signal-regulated kinase-1/2 signaling pathway activation. Mol Cancer. 2005:37. doi: 10.1186/1476-4598-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ, et al. MP1: A MEK Binding Partner That Enhances Enzymatic Activation of the MAP Kinase Cascade. Science. 1998:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 49.Boyd D. Regulation of Matrix Metalloproteinase Gene. J Cell Physiol. 2006:19–26. doi: 10.1002/jcp.20948. [DOI] [PubMed] [Google Scholar]
- 50.Wu JY, Lu H, Sun Y, Graham DY, Cheung HS, Yamaoka Y. Balance between Polyoma Enhancing Activator 3 and Activator Protein 1 Regulates Helicobacter pylori-Stimulated Matrix Metalloproteinase 1 Expression. Cancer Res. 2006:5111–5120. doi: 10.1158/0008-5472.CAN-06-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones LE, Humphreys MJ, Campbell F, Neoptolemos JP, Boyd MT. Comprehensive analysis of matrix metalloproteinase and tissue inhibitor expression in pancreatic cancer: increased expression of matrix metalloproteinase-7 predicts poor survival. Clin Cancer Res. 2004:2832–45. doi: 10.1158/1078-0432.ccr-1157-03. [DOI] [PubMed] [Google Scholar]
- 52.Shields MA, Dangi-Garimella S, Krantz SB, Bentrem DJ, Munshi HG. Pancreatic cancer cells respond to type I collagen by inducing snail expression to promote membrane type 1 matrix metalloproteinase-dependent collagen invasion. J Biol Chem. 2011:10495–504. doi: 10.1074/jbc.M110.195628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zucker S, Cao J. Selective matrix metalloproteinase (MMP) inhibitors in cancer therapy: Ready for prime time? Cancer Biol Ther. 2009:2371–2373. doi: 10.4161/cbt.8.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Utani A, Momota Y, Endo H, Kasuya Y, Beck K, Suzuki N, et al. Laminin α3 LGF4 Module Induces Matrix Metalloproteinase-1 through Mitogen-activated Protein Kinase Signaling. J Biol Chem. 2003:34483–34490. doi: 10.1074/jbc.M304827200. [DOI] [PubMed] [Google Scholar]
- 55.Durko M, Navab R, Shibata HR, Brodt P. Suppression of basement membrane type IV collagen degradation and cell invasion in human melanoma cells expressing an antisense RNA for MMP-1. Biochim Biophys Acta. 1997:271–80. doi: 10.1016/s0167-4889(97)00004-9. [DOI] [PubMed] [Google Scholar]
- 56.Chung L, Dinakarpandian D, Yoshida N, et al. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. The EMBO journal. 2004;23(15):3020–30. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pasternak RD, Hubbs SJ, Caccese RG, Marks RL, Conaty JM, DiPasquale G. Interleukin-1 stimulates the secretion of proteoglycan- and collagen-degrading proteases by rabbit articular chondrocytes. Clinical immunology and immunopathology. 1986;41(3):351–67. doi: 10.1016/0090-1229(86)90006-1. [DOI] [PubMed] [Google Scholar]
- 58.Poincloux R, Collin O, Lizarraga F, Romao M, Debray M, Piel M, et al. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. PNAS. 2011:1943–8. doi: 10.1073/pnas.1010396108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.