Abstract
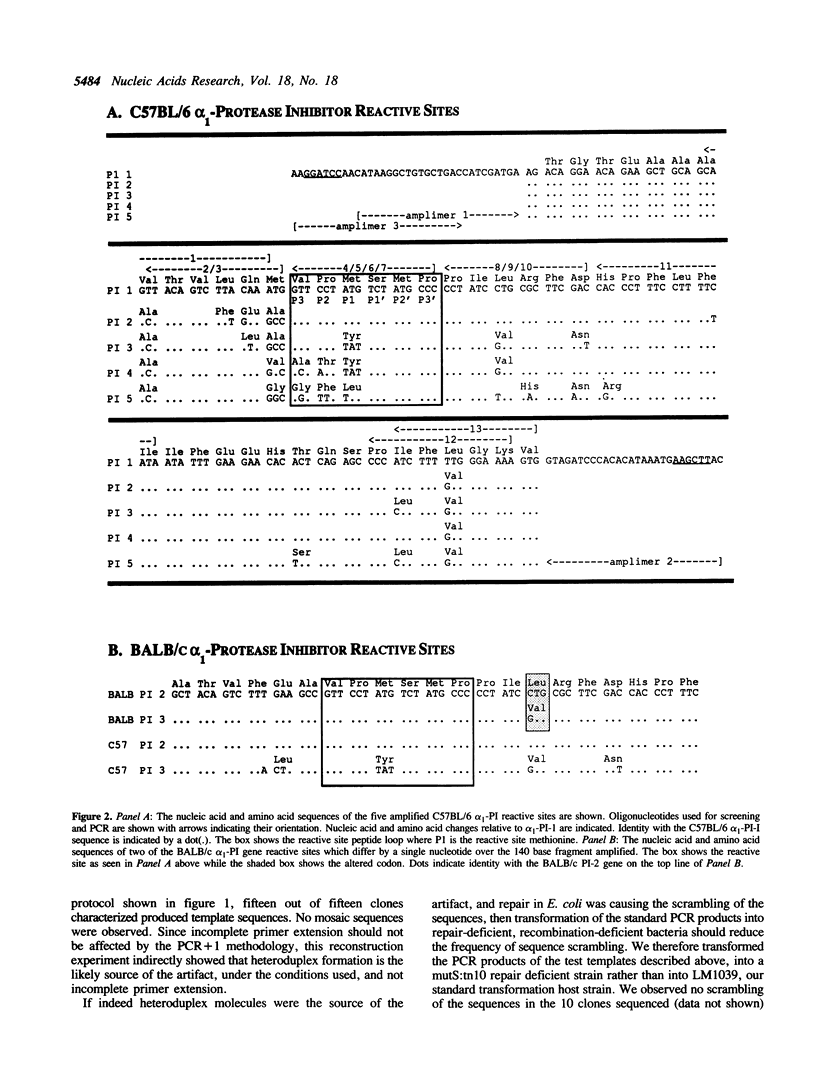
Murine protease inhibitor (alpha 1-PI) proteins are encoded by a multigene family which has undergone recent duplication. It has been suggested that the evolution of diversity within this gene family may be driven by unusual selection for novel function at the reactive site of the duplicated members (1,2,3). In an attempt to use polymerase chain reaction (PCR) to generate and sequence clones spanning the polymorphic reactive site region, a PCR artifact was identified and determined to result from heteroduplex formation during the co-amplification of the related sequences in this multigene system. This artifact results in sequences which are combinatorial mosaics of the template sequences. We present a simple and general method (PCR + 1) for overcoming this artifact and demonstrate its application in delineating five distinct alpha 1-PI reactive site sequences in C57BL/6 mice, thus providing sequence information to generate gene-specific probes. The significance of the reactive site diversity in this protease inhibitor gene family is discussed as well as the general applications and limitations of the PCR + 1 technique.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bao J. J., Sifers R. N., Kidd V. J., Ledley F. D., Woo S. L. Molecular evolution of serpins: homologous structure of the human alpha 1-antichymotrypsin and alpha 1-antitrypsin genes. Biochemistry. 1987 Dec 1;26(24):7755–7759. doi: 10.1021/bi00398a033. [DOI] [PubMed] [Google Scholar]
- Blank R. D., Campbell G. R., Calabro A., D'Eustachio P. A linkage map of mouse chromosome 12: localization of Igh and effects of sex and interference on recombination. Genetics. 1988 Dec;120(4):1073–1083. doi: 10.1093/genetics/120.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell R. W., Jeppsson J. O., Laurell C. B., Brennan S. O., Owen M. C., Vaughan L., Boswell D. R. Structure and variation of human alpha 1-antitrypsin. Nature. 1982 Jul 22;298(5872):329–334. doi: 10.1038/298329a0. [DOI] [PubMed] [Google Scholar]
- Carrell R. W., Pemberton P. A., Boswell D. R. The serpins: evolution and adaptation in a family of protease inhibitors. Cold Spring Harb Symp Quant Biol. 1987;52:527–535. doi: 10.1101/sqb.1987.052.01.060. [DOI] [PubMed] [Google Scholar]
- Cohen A. B. Mechanism of action of alpha-1-antitrypsin. J Biol Chem. 1973 Oct 25;248(20):7055–7059. [PubMed] [Google Scholar]
- Hill R. E., Hastie N. D. Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature. 1987 Mar 5;326(6108):96–99. doi: 10.1038/326096a0. [DOI] [PubMed] [Google Scholar]
- Hill R. E., Shaw P. H., Boyd P. A., Baumann H., Hastie N. D. Plasma protease inhibitors in mouse and man: divergence within the reactive centre regions. Nature. 1984 Sep 13;311(5982):175–177. doi: 10.1038/311175a0. [DOI] [PubMed] [Google Scholar]
- Johnson D., Travis J. Structural evidence for methionine at the reactive site of human alpha-1-proteinase inhibitor. J Biol Chem. 1978 Oct 25;253(20):7142–7144. [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Krauter K. S., Citron B. A., Hsu M. T., Powell D., Darnell J. E., Jr Isolation and characterization of the alpha 1-antitrypsin gene of mice. DNA. 1986 Feb;5(1):29–36. doi: 10.1089/dna.1986.5.29. [DOI] [PubMed] [Google Scholar]
- Laskowski M., Jr, Kato I., Kohr W. J., Park S. J., Tashiro M., Whatley H. E. Positive darwinian selection in evolution of protein inhibitors of serine proteinases. Cold Spring Harb Symp Quant Biol. 1987;52:545–553. doi: 10.1101/sqb.1987.052.01.062. [DOI] [PubMed] [Google Scholar]
- Laskowski M., Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Takahara H., Sinohara H. Inhibitory spectrum of mouse contrapsin and alpha-1-antitrypsin against mouse serine proteases. J Biochem. 1983 May;93(5):1411–1419. doi: 10.1093/oxfordjournals.jbchem.a134276. [DOI] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Travis J., Salvesen G. S. Human plasma proteinase inhibitors. Annu Rev Biochem. 1983;52:655–709. doi: 10.1146/annurev.bi.52.070183.003255. [DOI] [PubMed] [Google Scholar]
