Abstract
RAF inhibitors such as vemurafenib and dabrafenib block B-RAF-mediated cell proliferation and achieve meaningful clinical benefit in the vast majority of patients with B-RAFV600E-mutant melanoma. However, some patients do not respond to this regimen, and nearly all progress to therapeutic resistance. We employed a pooled RNA interference screen targeting >16,500 genes to discover loss of function events that could drive resistance to RAF inhibition. The highest-ranking gene was NF1, which encodes neurofibromin, a tumor suppressor that inhibits RAS activity. NF1 loss mediates resistance to RAF and MEK inhibitors through sustained MAPK pathway activation. However, cells lacking NF1 retained sensitivity to the irreversible RAF inhibitor AZ628 and an ERK inhibitor. NF1 mutations were observed in B-RAF-mutant tumor cells that are intrinsically resistant to RAF inhibition and in melanoma tumors obtained from patients exhibiting resistance to vemurafenib, thus demonstrating the clinical potential for NF1-driven resistance to RAF/MEK-targeted therapies.
Keywords: B-RAF, NF1, vemurafenib, PLX4720, resistance
Introduction
Cancer therapy has arguably entered a transformation fueled by the success of targeted agents such as kinase inhibitors deployed against tumors harboring “druggable” oncogene mutations. Unfortunately, resistance to these therapies remains a formidable challenge (1). In some cases of therapeutic resistance, cancer patients fail to respond to treatment at the outset (termed ‘de novo’ or innate resistance). Alternatively, resistance may emerge after several weeks or months of clinical response (termed ‘acquired’ resistance).
The identification of B-RAF mutations as key driver events in malignant melanoma spurred the development of small molecule inhibitors of the mitogen-activated protein kinase (MAP kinase) pathway in an effort to block dysregulated signal transduction engendered by the mutant B-RAF oncoprotein. As a result, B-RAF inhibitors such as vemurafenib or dabrafenib, or MEK inhibitors such as trametinib, elicited striking clinical response rates when administered as single agents in patients with B-RAFV600E-mutant melanomas (2–4). The use of B-RAF and MEK inhibitors in combination further extends the magnitude and duration of clinical benefit (5). However, intrinsic or acquired resistance to these regimens remains a major clinical problem. Systematic characterization of resistance to these agents is therefore needed in order to further the development of combined therapeutic strategies that either complement existing therapies or provide alternative treatment avenues.
Several mechanisms of resistance to vemurafenib have been described, most of which involve reactivation of downstream MEK/ERK signaling. Interestingly, secondary mutations involving the B-RAF gatekeeper residue (a threonine at codon 529) – common in drug-resistant CML and EGFR-mutant lung cancers – have not been observed, although preclinical data may support such a mechanism (6–9). Multiple laboratories have generated resistant cell line subclones by chronic exposure to RAF inhibitors in vitro, which may facilitate identification of resistance mechanisms by functional or genomic characterization of these cells. Such efforts have implicated amplification or mutation of RAS isoforms (10), enhanced C-RAF expression (11), activation of receptor tyrosine kinases (12) and a splice variant of B-RAF that constitutively dimerizes in the presence of inhibitor, producing sustained MEK/ERK signaling (13). Additionally, systematic gain of function screens identified COT (MAP3K8) as a resistance effector (14). Moreover, secretion of HGF by stromal cells may activate MET to promote resistance to B-RAF inhibition (15, 16). In the aggregate, these studies provided a rationale for the combined use of RAF and MEK inhibitors, and potentially additional therapeutic avenues that may overcome specific resistance mechanisms.
Genome-scale RNAi interference screens may offer a systematic and unbiased genetic approach to study tumor biology and therapeutic resistance. In order to elucidate potential loss-of-function mechanisms of resistance to B-RAF or MEK inhibition, we performed a pooled, lentiviral short hairpin RNA (shRNA) screen in drug-sensitive B-RAF-mutant melanoma cells exposed to high RAF inhibitor concentrations. Characterization of surviving cells identified inactivation of NF1 as a novel mechanism of resistance to both B-RAF and MEK inhibition.
Results
To identify genes capable of suppressing resistance to B-RAF inhibition, we utilized a library of 90,000 shRNAs targeting approximately 16,600 genes expressed in A375 cells, which harbor the B-RAFV600E mutation and are sensitive to small-molecule RAF and MEK inhibitors (17). Following infection with the lentiviral shRNA library, these cells were cultured in the presence of either DMSO or 3 μM of the RAF inhibitor PLX4720 for 14 and 7 population doublings respectively (Figure 1A and 1B). A drug-resistant population began to emerge within two weeks following shRNA infection (Figure 1B), whereas cells expressing a control shRNA targeting red fluorescent protein (shRFP) remained fully inhibited during this time frame (data not shown). The relative abundance of each shRNA was determined by PCR amplification and massively parallel sequencing of lentiviral library DNA (see Methods). The log fold-enrichment for shRNAs in the PLX4720-treated arm compared to the DMSO-treated arm was assessed. Using RNAi gene enrichment (RIGER) analysis (18), a ranked gene list was generated based upon the degree of enrichment of shRNAs targeting a given gene in the PLX4720-treated arm (Figure 1C). Only 31 genes had 2 or more shRNAs ranked in the top 1000 enriched subgroup (a criterion for candidacy as a “hit” from this screen)(Supplemental table 1). The top-ranking gene was NF1, which encodes the RAS-GTPase activating protein (RAS-GAP) neurofibromin (19). In particular, enrichment of 2 shRNAs targeting NF1 was clearly observed across six PLX4720-treated replicates (Figure 1D). This data raised the possibility that suppression of NF1 activity might permit proliferation of B-RAF-mutant melanoma cells in the presence of inhibitory RAF inhibitor concentrations.
Figure 1. A genome-wide RNAi pooled screen identifies NF1 as a key determinant of B-RAF inhibitor sensitivity in melanoma cells.
(A) Outline of the pooled screening strategy employed in B-RAFV600E A375 melanoma cells. A 90,000 shRNA pooled lentiviral library targeting approximately 16,600 genes was introduced into the cells, selection of infected cells was achieved using 0.5 μg/ml puromycin for 3 d. Cells were then treated with either DMSO or 3 μM PLX4720 for up to 14 population doublings. shRNA sequences were amplified by PCR and the relative abundance of each shRNA determined by Illumina sequencing.
(B) Growth of A375 cells infected with approximately 90,000 shRNAs and cultured in the presence of either DMSO or 3 μM PLX4720 for up to 14 population doublings.
(C) The number of reads per shRNA was normalized and log2 transformed and shRNA data for 2 DMSO controls and 6 PLX4720-treated replicas was analyzed using a ‘2nd best shRNA’, 2-class comparison of log-fold change (LFC) in RIGER to generate a ranked list of genes that were enriched in the PLX4720-treated cells. The screening hits are visualized by plotting the function y=1/normalized enrichment score (NES). The top 5-ranking candidate genes are indicated.
(D) Heat map of shRNA representation across early time point, DMSO-and PLX4720-treated replicas for the screen. shRNAs 39714 and 39717 were enriched across all drug-treated replicates.
(E) Validation of shRNA-mediated knockdown of NF1 in A375 cells. A375 cells were infected with shRNAs against luciferase or NF1, after selection in puromycin, cell lysates were made and levels of NF1 protein determined by Western blotting.
(F) A375 cells infected with either shLuc or shNF1 were cultured in the presence or absence of 3 μM PLX4720 for 2 weeks. Cells were fixed, then stained with crystal violet and photographed.
To validate this observation, A375 cells were re-infected with two distinct shRNAs targeting NF1. Reduced expression of NF1 protein was then confirmed by Western blotting (Figure 1E), and cell proliferation was assessed in the presence or absence of 3 μM PLX4720 for 2 weeks. Both NF1 shRNAs permitted robust cell proliferation (as measured by a colony formation assay in vitro) in the presence of PLX4720 (Figure 1F). These results suggested that loss of NF1 in A375 cells may reduce cellular dependency on oncogenic B-RAF for proliferation.
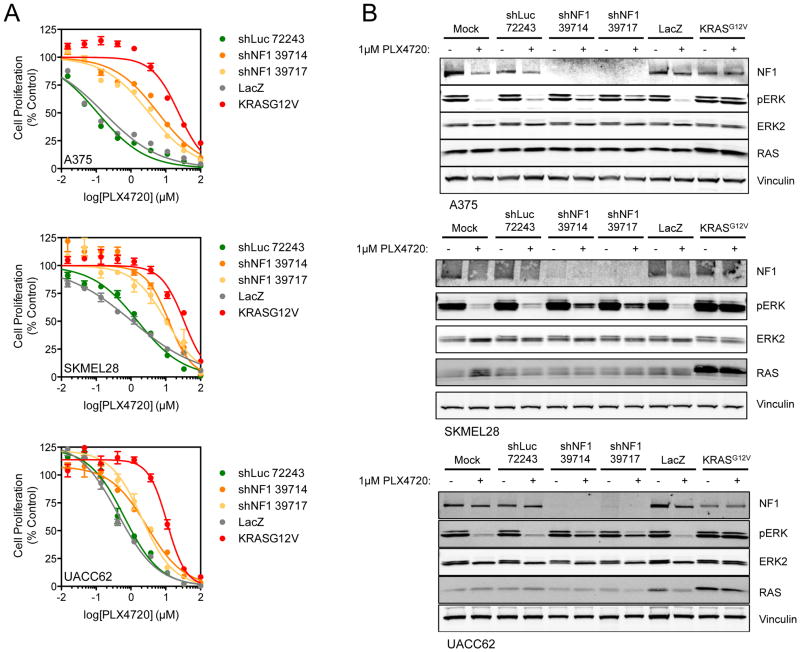
To determine whether suppression of NF1 could provide a generalizable loss-of-function mechanism of resistance to RAF inhibition, the B-RAF mutant melanoma cell lines SKMEL28 and UACC62 were treated with PLX4720 alongside A375 cells following infection with the shNF1 constructs. In each case, sustained cell proliferation in the presence of the RAF inhibitor was observed following NF1 silencing, compared to cells expressing a control shRNA (Figure 2A). Indeed, suppression of NF1 conferred a 5- to 31-fold shift in the GI50 values observed for PLX4720 (e.g. up to 2.2–3.5 μM compared to 0.112 μM in control A375 cells). By comparison, ectopic expression of oncogenic KRASG12V, a positive control for resistance to PLX4720 (and which is functionally equivalent to oncogenic forms of NRAS) (14) produced a GI50 of 26.3 μM. In parallel, cells were treated with 1 μM PLX4720 for 16 h and cell lysates were analyzed by Western blotting for ERK phosphorylation (Figure 2B). Loss of NF1 enabled sustained ERK phosphorylation in the presence of PLX4720 compared to mock-treated or shLuc-treated cells, although to a lesser extent than the KRASG12V control (Figure 2B). Thus, NF1 silencing was sufficient to confer resistance to RAF inhibition in multiple B-RAFV600E melanoma cell contexts.
Figure 2. Knockdown of NF1 drives resistance to selective B-RAF inhibitors through reactivation of the MAPK pathway.
(A) A375, SKMEL28 and UACC62 cells are resistant to PLX4720 following knockdown of NF1. shRNA-infected cells were treated with a 10-point concentration response of the inhibitor for 4 d and cell proliferation determined using cell-titer glo. Cells were also infected with LacZ and KRASG12V-expressing lentivirus to act as negative and positive controls respectively.
(B) Loss of NF1 is permissive for MAPK-signaling in the presence of B-RAF inhibitors. shRNA-infected A375, SKMEL28 and UACC62 cells were treated with the indicated concentrations of the inhibitors for 16 h, cell lysates were made and analyzed by Western blotting for the indicated proteins. Cells were also infected with LacZ and KRASG12V-expressing lentivirus to act as negative and positive controls respectively.
Recent whole-exome sequencing studies of melanoma tumors suggest that mutant NF1 might provide a “driver” genetic event that dysregulates MAP kinase signaling in some melanoma cells that lack B-RAF and NRAS mutations. B-RAF/NRAS “wild-type” melanoma cells are typically unresponsive to RAF inhibition (20–22). We therefore sought to test the ability of NF1 silencing to compensate for mutated B-RAF and modulate sensitivity to RAF inhibitors in an immortalized melanocyte model system (23). Knockdown of NF1 in primary human melanocytes expressing oncogenic B-RAF caused a 10-fold shift in the PLX4720 GI50, permitted robust proliferation in the presence of 3 μM PLX4720, and allowed sustained ERK phosphorylation in the presence of 0.2 and 1 μM PLX4720 (Supplemental Figure 1A–C). These experiments provided independent evidence that silencing of NF1 could confer resistance to RAF inhibition in a MAP kinase pathway-dependent manner.
Given that NF1 is a known negative regulator of RAS activity (24), we queried the activation state of RAS in A375 cells following NF1 knockdown using a RAS-GTP pull down assay. As expected, NF1 suppression caused a substantial increase in the level of active GTP-bound RAS (Figure 3A). Associated with the increased RAS-GTP, we observed a concomitant increase in C-RAF activation, as measured by phosphorylation of Ser338 (Figure 3B). C-RAF (Ser338) phosphorylation was further enhanced in the presence of PLX4720, suggesting that the enhanced RAS-GTP produced by NF1 silencing was competent to potentiate so-called “paradoxical” RAF activation (25–27).
Figure 3. Activation of RAS and C-RAF drives resistance to PLX4720.
(A) A375 cells were depleted of NF1 using shRNA and RAS-GTP levels in A375 cells were determined by a RAS-GTP affinity pull-down, followed by Western blotting for the indicated proteins.
(B) Combinatorial knockdown of NF1 and C-RAF abrogates NF1-mediated resistance to B-RAF inhibition at the level of ERK phosphorylation. A375 cells were infected with NF1 shRNA and treated with either DMSO or PLX4720 for 16 h. Cell lysates were analyzed for the indicated proteins.
(C) Combinatorial knockdown of NF1 and C-RAF abrogates NF1-mediated resistance to RAF inhibition. Quantitative analysis of the Western blots from Figure 3B for phospho-ERK normalized to ERK2 (red) and for cyclin D1 normalized to vinculin (green). Data from 3 independent experiments is presented.
(D) Combinatorial knockdown of NF1 and C-RAF abrogates NF1-mediated resistance to RAF inhibition. shRNA-infected cells were treated with a 10-point concentration response of the inhibitors for 4 d and cell proliferation determined using cell-titer glo.
Based on these observations, we next tested the requirement for C-RAF to mediate NF1-driven resistance to RAF inhibition by performing combined shRNA-mediated knockdown of both C-RAF and NF1, followed by assessment of MAPK signaling and sensitivity to RAF inhibition. Silencing of C-RAF alone had little effect on the responsiveness of MAPK signaling to PLX4720—this was expected given prior observations that C-RAF is inactive in B-RAFV600E melanoma under steady state conditions (28). However, under conditions of combined C-RAF/NF1 knockdown, ERK phosphorylation was inhibited more effectively compared to knockdown of NF1 alone (Figure 3B). Moreover, whereas cyclin D1 levels were inhibited by PLX4720 in A375 cells (as expected given the known regulation of this protein by MAP kinase signaling in B-RAFV600E melanoma), but NF1 silencing partially alleviated this effect (Figure 3B). Effects of PLX4720 on cyclin D1 protein expression were restored when both NF1 and C-RAF were depleted concomitantly. Quantitative analysis of the Western blots confirmed these observations (Figure 3C). Cellular sensitivity to PLX4720 followed a similar trend, whereby combinatorial knockdown of C-RAF and NF1 reversed the resistance to PLX4720 conferred by NF1 loss (Figure 3D).
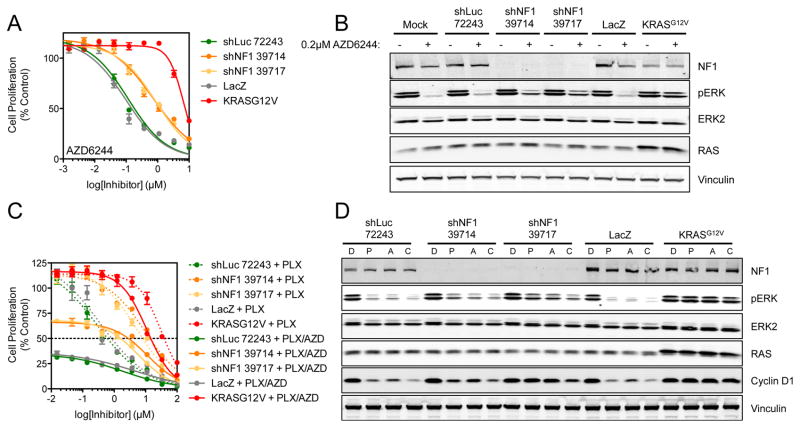
Having established that loss of NF1 could promote resistance to RAF inhibition, we sought to determine if NF1 silencing could also affect sensitivity to pharmacologic MEK inhibition. In particular, we hypothesized that inhibition of MEK, a substrate of both B-RAF and C-RAF, might have equal potency regardless of NF1 expression (given that MEK is a limiting MAPK kinase effector downstream of C-RAF). Interestingly, cells exhibiting NF1 knockdown were partially resistant to AZD6244 (Figure 4A). Here, NF1 silencing increased theGI50 to the MEK inhibitor AZD6244 by approximately 7-fold. ERK phosphorylation was also sustained in the presence of AZD6244 (Figure 4B). Combined inhibition of B-RAF and MEK by simultaneous treatment with PLX4720 and AZD6244 achieved greater efficacy than either agent alone; however, NF1 knockdown was still associated with residual resistance to combined RAF/MEK inhibition in vitro (Figure 4C). Moreover, a clear correlation was evident between residual ERK phosphorylation and inhibition of cell proliferation (Figure 4D). Nonetheless, a robust resistance phenotype remained apparent in the setting of NF1 knockdown, even under conditions of combined RAF/MEK inhibitor exposure.
Figure 4. Both MEK inhibition and combined RAF/MEK inhibition does not completely reverse NF1-mediated resistance.
(A) A375 cells infected with shRNAs targeting NF1 were treated with a 10-point concentration response of AZD6244 for 4 d and cell proliferation determined using cell-titer glo. Cells were also infected with LacZ and KRASG12V-expressing lentivirus to act as negative and positive controls respectively.
(B) In parallel to the above, cells were treated with 0.2 μM AZD6244 for 16 h and cell lysates analyzed by Western blotting for the indicated proteins.
(C) Combinatorial inhibition of B-RAF and MEK using PLX4720 and AZD6244 does not fully overcome NF1-mediated resistance. A375 cells were infected with the stated shRNAs/ORFs and then exposed to a 10-point concentration response of PLX4720 alone or PLX4720 plus 0.2 μM AZD6244 for 4 d. Cell proliferation was determined by cell-titer glo.
(D) Combined B-RAF/MEK inhibition does not fully block NF1-mediated resistance at the level of ERK phosphorylation. A375 cells were infected with shRNAs/ORFs as above and then treated with either DMSO (D), 1 μM PLX4720 (P), 0.2 μM AZD6244 (A) or a combination of PLX4720 and AZD6244 (C) for 16 h. Cell lysates were analyzed by Western blotting for the indicated proteins.
In the presence of activated RAS, PLX4720 can activate C-RAF through loss of feedback inhibition and induction of RAF-dimerization (25–27). Conceivably then, the RAS activation engendered by NF1 loss might promote a biochemical state wherein the activated RAF/MEK complex is less vulnerable to MEK inhibition, particularly under conditions of concomitant RAF inhibitor exposure. To test this hypothesis, we used the irreversible RAF inhibitor AZ628. This compound prevents C-RAF activation due to persistent occupation of the ATP binding site (25). In contrast to the aforementioned drug conditions, only minimal resistance (2-fold) was observed to AZ628 (Figure 5A). Moreover, ERK phosphorylation was effectively suppressed (between 80–90%) by this irreversible inhibitor, even in the setting of NF1 knockdown but only by 65% in the presence of oncogenic KRAS (Figure 5B). Furthermore, combined NF1/C-RAF knockdown resensitized cells to treatment with the MEK inhibitor (Supplemental Figure 2). These results provided additional support for the notion that C-RAF signaling is required for the resistance phenotype induced by NF1 loss.
Figure 5. Cells lacking NF1 expression are sensitive to sustained B-RAF/C-RAF inhibition and to ERK inhibition.
(A) A375 cells infected with the indicated shRNAs/ORFs were treated with a 10-point concentration response of AZ628 for 4 d and cell proliferation determined using cell-titer glo.
(B) In parallel to the above, cells were also treated with 0.2 μM AZ628 for 16 h, cell lysates were analyzed by Western blotting for the indicated proteins.
(C) A375 cells infected with the indicated shRNAs/ORFs were treated with a 10-point concentration response of VTX-11e for 4 d and cell proliferation determined using cell-titer glo.
(D) In parallel to the above, cells were also treated with 1μM VTX-11e for 16 h, cell lysates were analyzed by Western blotting for the indicated proteins.
We also tested the effect of pharmacologic ERK inhibition in this setting using the compound VTX-11e (29). Interestingly, B-RAFV600E melanoma cells harboring NF1 knockdown remained sensitive to ERK inhibition (Figure 5C). Consistent with this observation, phosphorylation of the ERK substrate FRA1 was inhibited equally in the presence or absence of NF1 knockdown (Figure 5D). This pattern of activity was confirmed in colony formation assays, whereby robust resistance was observed to the selective RAF inhibitors PLX4720, PLX4032, GDC0879 and the MEK inhibitor AZD6244. However, NF1 knockdown produced only modest resistance to the irreversible RAF inhibitor AZ628, and no resistance to the ERK inhibitor VTX-11e (Supplemental Figure 3A&B).
To understand the potential for endogenous NF1 inactivation to mediate resistance to RAF inhibition, we leveraged the Cancer Cell Line Encyclopedia (CCLE) (30) to identify cell line models that contained putative inactivating NF1 mutations in the context of oncogenic B-RAFV600E mutation. Toward this end, over 500 CCLE lines have previously been profiled for pharmacologic sensitivity to RAF and MEK inhibition (30). We identified five B-RAFV600E cell lines (3 melanomas and 2 colorectal tumors) with co-occurring mutations in NF1 (Supplemental Figure 4). Of these, three contained obvious “damaging” NF1 mutations (HS695T: NF1Q959*, LOXIMVI: NF1Q1174*and RKO: NF1L626fs,Q2340fs,V2205A) whereas two contained point mutations (WM88: NF1R1306Q, LS411N: NF1T2805I), deemed unlikely to affect NF1 function according to the Mutation Assessor algorithm (data not shown)(31). HS695T and RKO cells also exhibited reduced NF1 mRNA expression. Firstly, we assessed NF1 protein expression by Western blotting and those cell lines with damaging mutations in NF1 expressed significantly less protein than the wild-type cells (Figure 6A). The sensitivity of HS695T, LOXIMVI and RKO cells to MAPK pathway inhibition by PLX4720 was assessed alongside A375, SKMEL28 and UACC62 B-RAFV600E/NF1WT melanoma cells. B-RAFV600E cell lines that expressed no or undetectable levels of NF1 protein (LOXIMVI and RKO) were highly resistant to the inhibition of ERK phosphorylation by PLX4720, whereas HS695T cells which express low levels of NF1 protein were still relatively sensitive (Figure 6B). In accord with this finding, NF1 mutation correlated with resistance to PLX4720 in these cells (Figure 6C). The mean GI50 was 0.247 μM in NF1-wild-type cells which increased greater than 50-fold in the NF1-mutant cell lines to 15.8 μM
Figure 6. Loss of NF1 is observed in some B-RAF-mutant human melanoma and colorectal cell lines.
(A) Melanoma and colorectal cancer cell lines from the Broad Cancer Cell Line Encyclopedia were assessed for expression of B-RAFV600E and damaging mutations in NF1. 3 such lines were identified (HS695T, LOXIMVI and RKO, colored red) and analyzed alongside a panel of B-RAFV600E/NF1WT cells (colored blue) for expression of NF1 protein by quantitative Western blotting. A two-tailed t-test was performed to determine if the expression of NF1 protein was significantly different between cells expressing either wild-type or mutant protein. Incomplete suppression of ERK phosphorylation by PLX4720 in NF1-mutant cell lines. Cells were treated with the indicated concentrations of PLX4720 for 16 h, cells were analyzed for ERK2 and phospho-ERK1/2 by In-cell Western. Cells with wild-type NF1 are labeled blue, those with mutant NF1 are labeled red.
(B) Reduced expression of NF1 protein is associated with resistance to PLX4720. NF1-wild-type and mutant cell lines were treated with a 10-point titration of PLX4720 for 16 h and ERK phosphorylation was assessed using an in-cell Western assay. Cells with wild-type NF1 are labeled blue, those with mutant NF1 are labeled red.
(C) Reduced expression of NF1 protein is associated with resistance to PLX4720. NF1-wild-type and mutant cell lines were treated with a 10-point titration of PLX4720 for 4 d and cell proliferation was assessed using cell-titer glo. Cells with wild-type NF1 are labeled blue, those with mutant NF1 are labeled red.
(D) Human B-RAF- and NF1-mutant melanoma and colorectal cancer cell lines display only modest resistance to inhibition of ERK. Cells were exposed to a 10-point titration of the ERK inhibitor VTX-11e for 4 d and cell proliferation was assessed using cell-titer glo. Cells with wild-type NF1 are labeled blue, those with mutant NF1 are labeled red.
Given that NF1 knockdown did not confer resistance to pharmacologic ERK inhibition in A375 cells (as described above), we tested the hypothesis that B-RAFV600E/NF1-mutant cancer cell lines might remain sensitive to VTX-11e. Notably, the ERK inhibitor showed activity against NF1-mutant cell lines with a mean GI50 of 0.781 μM compared to 0.219 μM in B-RAF-mutant cells with wild-type NF1 (Figure 6D). These results suggested that pharmacologic ERK inhibition might in principle provide a viable therapeutic strategy in B-RAFV600E cancer cells harboring NF1 loss.
To confirm that NF1 alterations may confer resistance to B-RAF inhibition in human melanoma, we assessed the status of NF1 using whole exome sequencing of tumors from patients who had relapsed during vemurafenib treatment. We identified four patients whose tumors expressed mutant alleles of NF1 (Figure 7A). Of these, one was a nonsense mutation (patient 46, p.R2450*) that would likely result in reduced protein expression. This mutation was present in both pre-treatment and post-relapse biopsies, and interestingly the patient had a progression-free survival of only ~10 weeks. This short duration of response raised the possibility that the concomitant NF1 mutation conferred intrinsic resistance to vemurafenib (Figure 7B). The other NF1 mutations were silent events (patient 15, c.135T>C, patient 45 c.4023G>A and patient 50 c3018C>T) that are not predicted to alter NF1 protein expression. However, given that NF1 is known to be affected by splice-site mutations (32), we assessed the predicted impact of these mutations on both canonical splice sites and exon splice enhancer sites (ESEs) using the Human Splicing Finder (33). This analysis revealed that all three mutations were located within candidate splicing motifs. Conceivably, then, these mutations may produce aberrant NF1 exon splicing. Like patient 46, patients 15 and 50 exhibited response rates of relatively short duration (10 and 12 weeks, respectively). Moreover, these mutations were also observed in both the pre-treatment and post-relapse biopsies, consistent with intrinsic resistance to vemurafenib. In contrast, patient 45 experienced a progression-free survival of five months, and the NF1 mutation was only present in the post-relapse biopsy (Figure 7C), indicative of acquired resistance. Together, these observations provide preliminary support for the notion that genomic dysregulation of NF1 may influence both de novo and acquired resistance to RAF inhibition in the clinical arena.
Figure 7. Whole-exome sequencing identifies NF1 mutations in melanoma patient tumors exhibiting resistance to vemurafenib.
(A) Melanoma patients who progressed on treatment with vemurafenib were biopsied pre- and post-treatment. Whole-exome sequencing was performed and NF1 alterations assessed. The NF1 mutations are listed alongside the functional impact on the expressed protein. Silent mutations were assessed for their potential effects on splicing sites using the human splicing finder. The splicing motif affected by the mutation (in red) is indicated and ‘site broken’ indicates potentially damaging effects on splice site function.
(B) Progression free survival data was extrapolated from Chapman et al (2) and the number of patients who progressed per month is in blue, the cumulative number of progressing patients is in gray. NF1 mutations observed in our cohort of patients are overlaid at their respective PFS times. Patients with a PFS of less than three months were nominated for de novo resistance, while those with a PFS of greater than or equal to three months demonstrated acquired resistance. Three NF1 alterations are observed in pre-treatment B-RAF-mutant melanoma patient samples, and all three patients demonstrated de novo resistance. A fourth patient with a PFS of five months exhibited acquired resistance.
(C) Integrative Genomics Viewer (48) plot showing an NF1 silent mutation observed in the post-relapse biopsy sample only (c.4023G>A) in a patient with a PFS of five months.
Discussion
In malignant melanoma, activating B-RAF mutations confer exquisite dependency on RAF/MEK/ERK signaling, which has been successfully targeted using small molecule RAF and MEK inhibitors. However, therapeutic resistance inevitably develops following a period of disease stabilization or regression, and some B-RAFV600E-mutant melanomas exhibit intrinsic resistance to these agents (2). Understanding the mechanistic basis of drug resistance is essential to the development of new therapeutic strategies that maximize clinical benefit. Here, we queried resistance to RAF inhibition using an unbiased, genome-scale RNA interference screen for modifiers of sensitivity to small molecule RAF inhibition. Our approach builds upon complementary studies using gain-of-function approaches, such as systematic open reading frame (ORF) overexpression screens, (14) or the generation of resistant cell lines by chronic exposure to inhibitors (10, 11, 13, 34).
The identification of NF1 loss as a resistance effector describes one of the first loss-of-function events capable of mediating resistance to RAF inhibitors identified by systematic functional approaches. The protein encoded by the NF1 gene, neurofibromin, is a known tumor suppressor gene and negative regulator of RAS proteins. This function is achieved through stimulation of the GTPase activity of RAS by neurofibromin, thus converting it from an active, GTP-bound form to its inactive GDP-bound form. Therefore, loss of NF1 activates RAS and provides an upstream stimulus to activate C-RAF, driving resistance to a RAF inhibitor by reactivation of the MAPK pathway. This is consistent with other reported mechanisms whereby RAS is mutated or amplified in vemurafenib-resistant cell lines, signaling through C-RAF to maintain MAPK signaling (10, 35). These findings are therefore consistent with several published reports indicating that the majority of resistance mechanisms reported for RAF inhibitors involve reactivation of the MAPK pathway. This underscores a fundamental dependency on RAS signaling potentiated by NF1 loss. This event also confers a C-RAF dependency in the setting of pharmacologic RAF inhibition, as evidenced by a near-complete reversal of the resistance phenotype by C-RAF knockdown. This observation raises the possibility that, in the future, saturating pharmacologic inhibition of both B- and C-RAF (and possibly A-RAF as well) could reduce the capacity for resistance to develop. Such inhibition might be achieved by an irreversible RAF inhibitor that both promotes an inactive conformation of RAF. Consistent with this notion is the observation that NF1 was unable to confer resistance to AZ628, a tool compound with these characteristics (25). Consequently, we hypothesized that either a single-agent MEK inhibitor or combined RAF and MEK inhibition might circumvent NF1-mediated resistance. Indeed, combined exposure displayed more potent activity in vitro than did either agent alone in the setting of NF1 knockdown. However, the drug combinations tested herein did not fully restore sensitivity, at least in vitro. Conceivably, this limitation may be overcome by more potent MEK inhibitors or clinical RAF/MEK regimens that enable more complete suppression of MEK/ERK signaling. Alternatively, C-RAF may have MEK-independent roles in tumor cell survival and proliferation (36–39). Therefore, whilst combined RAF/MEK inhibition certainly improves clinical responses in B-RAF-mutant melanoma (5), the potential for NF1 loss (and perhaps other mechanisms of upstream RAS-activation) to drive resistance may remain. On the other hand, melanoma cells harboring NF1 knockdown remained sensitive to a selective ERK inhibitor, suggesting that ERK may represent a pivotal integration point for upstream MAP kinase signaling, and may offer therapeutic potential for circumventing multiple resistance mechanisms.
To ascertain whether NF1 loss might provide an endogenous resistance mechanism, we analyzed cancer cell lines in which NF1 mutation co-occurred with B-RAFV600E mutation. Using the Cancer Cell Line Encyclopedia, we identified 5 B-RAFV600E cell lines that had NF1 mutations, 3 of which were nonsense mutations. Each of these cell lines was insensitive to PLX4720, consistent with the NF1 shRNA screening and validation results in RAF-inhibitor-sensitive cell lines. Furthermore, by integrating our findings with a published cohort of 121 primary melanoma tumors, metastatic tumors and short-term cultures (20) previously analyzed by whole exome sequencing, we gained additional insights into the potential clinical relevance of NF1 mutations. In this cohort, putatively damaging NF1 mutations were observed in 25% (5/21) of samples that lacked highly recurrent mutations in either B-RAF or NRAS. Genomic studies of an independent melanoma cohort have also observed this association (21). However, NF1 mutations were also observed in B-RAF- or NRAS-mutant tumors, including a splice site mutation (NF1G1166) co-occurring with the activating mutant B-RAFK601E,P731S, a nonsense mutation (NF1R440*) co-occurring with NRASQ61K, and a GAP-related domain mutation (NF1P1323L) in a B-RAFV600E tumor. Given that both melanoma and colorectal tumors harbor coexisting B-RAF and NF1 mutations, it therefore seemed likely that clinical scenarios could arise where NF1 loss serves as a gating resistance effector in the setting of B-RAF/MEK inhibition. Toward this end, we assessed NF1 alterations by whole exome sequencing in the tumors of melanoma patients treated with vemurafenib who progressed whilst on treatment. We identified four patients whose tumors had NF1 alterations that could impact upon protein function or expression. One event resulted in a nonsense mutation in NF1 (p.R2450*), likely to result in nonfunctional protein. The other three mutations were silent events but importantly, they occurred in putative splice regulatory or exon splicing enhancer sites predicted to regulate proper mRNA processing. These data affirm the potential for NF1 alterations to drive both de novo and acquired resistance to vemurafenib in melanoma. As deep genomic/molecular characterization accrues for tumor specimens obtained prior to treatment, during treatment and following relapse, the relevance of NF1 loss to clinical resistance should become clearer.
Our data may also endorse the development of irreversible RAF inhibitors. Such agents may in principle show benefit in NRAS-mutant tumors because of their enhanced dependency upon C-RAF signaling for tumor cell growth. Alternatively, the use of ERK inhibitors may overcome numerous melanoma resistance mechanisms if such agents can be administered safely at doses that achieve robust target inhibition. Toward this end, both irreversible-RAF and ERK inhibitors are entering preclinical/clinical trials, thereby offering forthcoming opportunities to evaluate their efficacy in B-RAF-mutant cancers. Altogether, these results demonstrate the promise of systematic preclinical studies combined with knowledge of tumor genetic/molecular alterations to elucidate cancer drug resistance mechanisms that may inform novel therapeutic avenues for many types of cancer.
Materials and Methods
Cell culture and reagents
Cell lines were obtained from the American Tissue Culture Collection (ATCC) or the National Cancer Institute (NCI) and cultured in either RMPI1640 or DMEM (Cellgro), 10% FBS (Gemini Bio-Products) as recommended by the suppliers. Cells were passaged for less than 6 months after receipt and authenticated by short tandem repeat profiling. PLX4720, PLX4032, AZ628, GDC0879 and AZD6244 were purchased from Selleck Chemicals and the ERK inhibitor VTX-11e was synthesized as previously reported (29). Expression constructs for KRASG12V have been described previously (40). Primary human melanocytes expressing Myc-tagged B-RAFV600E were cultured in Ham’s F10 medium supplemented with 10% FBS (23).
Pooled lentiviral shRNA screen
A375 cells were seeded into 12-well plates at a density of 3×106 cells/well, and a total of 7.2×107 cells/replicate were infected with a 90,000 shRNA pooled library at an MOI of 0.3–0.5 (18). Cells were centrifuged for 2 h at 2000 rpm, followed by an immediate medium change. The next day, cells were pooled and transduced cells were selected by culturing in the presence of 0.5 μg/ml puromycin for 3 d. Cells were then seeded into T225 flasks for each replica and treated with either DMSO or 3 μM PLX4720. DMSO-treated cells were passaged for up to 14 population doublings and PLX4720-treated cells were passaged for approximately 7 population doublings. Cells were harvested by trypsinization and stored at −80 °C in PBS. Genomic DNA was extracted as described previously (18) and the shRNA sequences were amplified by PCR (41). Here, 140 μg of genomic DNA was split over 60 reactions per sample, reactions were then re-pooled per sample prior to a secondary PCR step. Illumina adapters and independent sample barcodes were incorporated during this secondary PCR step using a scaled-up secondary PCR reaction consisting of six 100 μl reactions, followed by massively parallel sequencing (Illumina). The number of reads for each shRNA was incorporated into the following calculation to normalize between samples: Log2((shRNA reads/total reads for sample)×1e6). The enrichment of shRNAs in the PLX4720-treated arm relative to the DMSO arm was determined using the RIGER algorithm, ‘2nd-best shRNA’ analysis in GENEE (http://www.broadinstitute.org/cancer/software/GENE-E/)(18) to produce a ranked list of statistically significant genes based on the degree of corresponding shRNA enrichment in the PLX4720-resistant population. By ranking genes according to the ranking of the ‘2nd best shRNA’ per gene, results are influenced less by high-ranking, single shRNA hits for a given gene.
shRNA/ORF constructs and lentiviral infections
All shRNA expression constructs were obtained from The RNAi Consortium and the Broad Institute RNAi Platform. shRNAs were expressed from the lentiviral expression plasmid pLKO.1 or pLKO-TRC005, and virus was produced by transfection of approximately 2.4×106 293T cells with 3 μg pLKO.1, 2.7 μg Δ8.9 (gag, pol) and 0.3 μg VSV-G plasmids using 18 μl Fugene6 (Promega). Viral supernatant was harvested 72 h post-transfection. Mammalian cells were infected at a 1:100 dilution of virus in the presence of 4 μg/ml polybrene (Millipore) and centrifuged for 30 min at 2000 rpm. The culture medium was changed immediately after spin infection and 24 h later, puromycin at a concentration of 0.5–1 μg/ml was added to select for infected cells. pLX304 plasmids for LacZ and KRASG12V were used to generate lentivirus as above. Cells were infected in the same manner except 10 μg/ml blasticidin (Life Technologies) was used to select for infected cells.
shRNA constructs used
The shRNA constructs used in this study were:
Cell proliferation assays
Cells were seeded at a density of 0.5–5×103 cells/well in 96 well plates. The next day cells were treated with 4 μg/ml polybrene and transduced with shRNA-expressing lentivirus by centrifugation for 30 min at 2000 rpm followed by an immediate medium change. After 3 d the medium was again changed and compounds added to the required concentrations. After a further 4 d, cell proliferation was assessed using the cell-titer glo reagent (Promega). GI50 values were determined using GraphPad Prism. For colony formation assays, shRNA-infected cells were seeded at 300 cells/well of a 12-well plate and incubated with 3 μM PLX4720 or DMSO for 2 weeks. Medium containing either DMSO or PLX4720 was changed every 3–5 d. Cells were fixed with 4% formaldehyde and stained with 0.5% crystal violet; plates were then washed with distilled water and photographed.
Protein analysis
Cells were seeded at 5–10×104 cells/well in 6 well plates. Following treatment, cells were then washed with PBS and lysed in 1% SDS lysis buffer. Following protein normalization using the BCA reagent (Sigma), equal amounts of protein were resolved by SDS-PAGE (Life Technologies) and transferred to PVDF-Fl membranes (Bio-Rad). Western blots were blocked with Li-Cor blocking buffer and then probed with the desired antibodies overnight at 4 °C. Bands were detected using IR fluorescence and an Odyssey scanner (Li-Cor Biosciences). Antibodies were obtained from Cell Signaling, Santa Cruz Biotechnology, Sigma, Millipore, BD Biosciences, Li-Cor Biosciences and Thermo-Fisher Scientific. The determination of RAS-GTP levels in cell lysates was performed using the RAS Activation Assay Kit (Millipore).
In-cell Western
Cells were seeded in 96 well clear-bottomed, black micro titer plates at 2×104 cells/well. The next day, cells were treated with a 10-point titration of PLX4720 for 16 h. Cells were fixed in 4% formaldehyde, 0.1% TX-100 in phosphate-buffered saline for 30 min. The wells were blocked with Li-Cor blocking buffer for 30 min and incubated with antibodies to phospho-ERK (Sigma) and ERK2 (Santa Cruz Biotechnology) overnight at 4 °C. Cells were washed 3 times with 200 μl 0.1% tween 20 and incubated with Li-Cor secondary antibodies (anti-mouse IR800, anti-rabbit IR680) for 1 h. The plates were scanned using an Odyssey scanner (Li-Cor) and quantified using the manufacturer’s software. ERK phosphorylation was calculated as a percentage of vehicle-treated controls.
Clinical Samples
All melanoma and matched normal samples analyzed were collected and sequenced under an Institution Review Board approved protocol (COUHES#0806002814). Standard Broad Institute Sequencing Platform techniques were used for DNA extraction and quality assessment (20).
Library Preparation, Assembly, and Quality Control
Exome capture and library construction were performed as previously described (42), and libraries were sequenced on Illumina HiSeq 2000 machines. The resulting sequencing data obtained from the Illumina pipeline were assembled using the Picard pipeline (43). Cross-contamination of samples was estimated using ContEst (44), and samples with > 5% contamination were excluded from this study. SNP fingerprints from each lane of a tumor/normal pair were crosschecked to confirm concordance, and non-matching lanes were removed from analysis.
Identification of Somatic Mutations
Somatic single nucleotide base-pair substitutions were identified using MuTect (45). Annotation of identified mutations for mutation effect was done using Oncotator (46). These algorithms were executed using the Broad Firehose Infrastructure (47).
Supplementary Material
Significance.
This work identifies functional loss of NF1 as a mediator of resistance to RAF inhibitors in B-RAFV600E-mutant cancers. Furthermore, we nominate new therapeutic modalities to treat this mechanism of resistance.
Acknowledgments
Financial support: This study was supported by grants from the National Cancer Institute (5P50CA127003-05) STARR Cancer Consortium (I4-A448), the NIH New Innovator Award (DP2OD002750) and the Melanoma Research Alliance. JTP is supported by the Swiss Foundation for Grants in Biology and Medicine (PASMP3_134379/1).
Footnotes
Conflicts of interest: L.A.G. is a consultant for Foundation Medicine, Novartis, and Millennium/Takeda, and an equity holder in Foundation Medicine.
References
- 1.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–9. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 4.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK Inhibition in Melanoma with BRAF V600 Mutations. N Engl J Med. 2012 doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 7.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 8.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamborini E, Bonadiman L, Greco A, Albertini V, Negri T, Gronchi A, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–9. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–54. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 20.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci U S A. 2010;107:14903–8. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 24.Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, et al. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63:835–41. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 25.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 26.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–91. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 29.Aronov AM, Tang Q, Martinez-Botella G, Bemis GW, Cao J, Chen G, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J Med Chem. 2009;52:6362–8. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 30.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pros E, Gomez C, Martin T, Fabregas P, Serra E, Lazaro C. Nature and mRNA effect of 282 different NF1 point mutations: focus on splicing alterations. Hum Mutat. 2008;29:E173–93. doi: 10.1002/humu.20826. [DOI] [PubMed] [Google Scholar]
- 33.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su F, Bradley WD, Wang Q, Yang H, Xu L, Higgins B, et al. Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation. Cancer Res. 2012;72:969–78. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Fujii K, Zhang L, Roberts T, Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci U S A. 2001;98:7783–8. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mielgo A, Seguin L, Huang M, Camargo MF, Anand S, Franovic A, et al. A MEK-independent role for CRAF in mitosis and tumor progression. Nat Med. 2011;17:1641–5. doi: 10.1038/nm.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navas TA, Baldwin DT, Stewart TA. RIP2 is a Raf1-activated mitogen-activated protein kinase kinase. J Biol Chem. 1999;274:33684–90. doi: 10.1074/jbc.274.47.33684. [DOI] [PubMed] [Google Scholar]
- 39.O’Neill E, Rushworth L, Baccarini M, Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science. 2004;306:2267–70. doi: 10.1126/science.1103233. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–61. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashton JM, Balys M, Neering SJ, Hassane DC, Cowley G, Root DE, et al. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell Stem Cell. 2012;11:359–72. doi: 10.1016/j.stem.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.http://picard.sourceforge.net/
- 44.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.https://confluence.broadinstitute.org/display/CGATools/MuTect
- 46.https://confluence.broadinstitute.org/display/CGATools/Oncotator
- 47.https://confluence.broadinstitute.org/display/CGATools/Firehose
- 48.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.