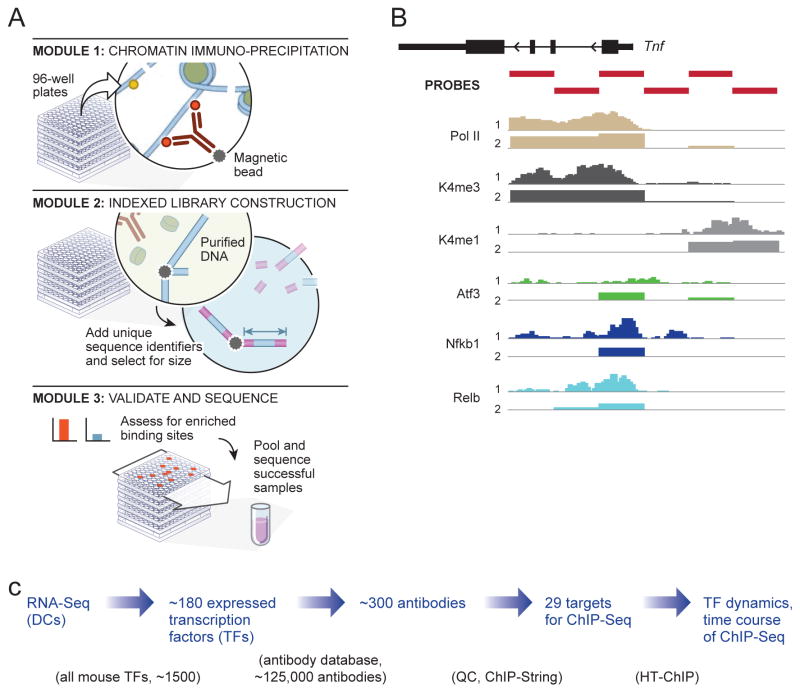
Figure 1. High throughput Chromatin Immuno-precipitation (HT-ChIP) pipeline.
A. Blueprint of the HT-ChIP pipeline Top, Protein-DNA fragments are precipitated using antibody coupled magnetic beads in 96-well plates. Middle. Precipitated DNA is purified using magnetic beads, indexed adapters are ligated and size selected to generate sequencing libraries. Bottom. Samples are validated using ChIP-String; successful samples are pooled and sequenced. B. ChIP-String validation. Nanostring probes (red) target selected active regulatory regions. Comparison of (a) ChIP-Seq (b) or ChIP-String for K4me1 (gray) K4me3 (dark grey), Pol-II (light brown) Relb, and Nfkb1 (variants of blue), and Atf3 (green) C. Strategy for ab initio TF-DNA binding maps. The strategy consists of four steps 1. Expression analysis using RNA-Seq (2) Selection of top expressed TFs (3) Screening for all potential ChIP-Seq antibodies (4) ChIP in appropriate time points all validated TF targets.