Abstract
Mitochondria are membrane-bound cytoplasmic organelles that serve as the major source of ATP production in eukaryotic cells. GABP (also known as nuclear respiratory factor 2) is a nuclear E26 transformation-specific transcription factor (ETS) that binds and activates mitochondrial genes that are required for electron transport and oxidative phosphorylation. We conditionally deleted Gabpa, the DNA-binding component of this transcription factor complex, from mouse embryonic fibroblasts (MEFs) to examine the role of Gabp in mitochondrial biogenesis, function, and gene expression. Gabpα loss modestly reduced mitochondrial mass, ATP production, oxygen consumption, and mitochondrial protein synthesis but did not alter mitochondrial morphology, membrane potential, apoptosis, or the expression of several genes that were previously reported to be GABP targets. However, the expression of Tfb1m, a methyltransferase that modifies ribosomal rRNA and is required for mitochondrial protein translation, was markedly reduced in Gabpα-null MEFs. We conclude that Gabp regulates Tfb1m expression and plays an essential, nonredundant role in mitochondrial biogenesis.
INTRODUCTION
Mitochondria are semiautonomous, membrane-bound cytoplasmic organelles that are the major source of ATP production in the eukaryotic cell. In addition to their roles in generating cellular energy through electron transfer and oxidative phosphorylation, mitochondria are also required for lipid biosynthesis, cell signaling, apoptosis, and other essential cellular functions. Because the 16-kb mitochondrial genome does not encode any transcription factors, it depends on nuclear transcription factors for the expression of mitochondrial DNA (mtDNA) genes. Mitochondrial transcription factor A (TFAM) and TFBM are nuclear transcription factors that exclusively regulate mtDNA genes. Other transcription factors, including NRF1 (nuclear respiratory factor 1) and NRF2, control both mtDNA genes and nuclear genes (1–4).
There are two distinct TFBM proteins, TFB1M and TFB2M, both of which have rRNA methyltransferase activity (5, 6). Shadel and colleagues reported that TFB1M and TFB2M play crucial but distinct roles in the control of mitochondrial biogenesis in both human and Drosophila cells (7–9). TFB2M mainly regulates mtDNA replication and mitochondrial gene transcription. Ectopic expression of TFB1M did not significantly affect mitochondrial function, but reduced expression of TFB1M decreased mitochondrial protein translation through impaired methylation of the 12S rRNA, impaired mitochondrial ribosome assembly, and abolished mitochondrial translation. Genetic disruption of Tfb1m caused early (embronic day 8.5 [E8.5]) embryonic lethality but neither activated nor repressed mitochondrial gene transcription (10). Thus, adequate levels of TFB1M are required for normal mitochondrial protein synthesis and biogenesis.
Scarpulla and colleagues recognized that the multiprotein NRF2 complex is the human homologue of GABP, or GA-binding protein (11). GABP is the only obligate multimer among the more than two dozen mammalian E26 transformation-specific (ETS) factors (12). The tetrameric GABP complex includes two distinct proteins; GABPα binds to DNA through its ETS domain, and it recruits GABPβ, which activates transcription through a glutamine-rich region in its carboxy terminus (13). GABPA is a unique gene in the human and murine genomes (14), and GABPα is the only protein that can recruit GABPβ to DNA to form the transcriptionally active complex (15). GABP regulates lineage-restricted myeloid and lymphoid genes that are required for innate immunity (13), is required for cell cycle control in fibroblasts (16), and has been implicated in the regulation of more than one dozen mitochondrial genes (2). Genetic disruption (16, 17) and knockdown (18) of mouse Gabpa caused early embryonic lethality, and mitochondrial dysfunction was proposed as a possible explanation for embryonic loss (17). However, it has been unclear if GABP, as a member of the large ETS family of transcription factors that bind to similar DNA motifs, is an essential, nonredundant regulator of mitochondrial gene expression.
To examine the role of GABP in the regulation of mitochondrial biogenesis, function, and gene expression, we conditionally deleted Gabpa from cultured primary mouse embryonic fibroblasts (MEFs). Mitochondrial mass in Gabpα-null MEFs was reduced by approximately one-third, and ATP production, oxygen consumption, and mitochondrial protein were decreased proportionally; however, mitochondrial structure, membrane potential, and apoptosis were not significantly altered by loss of Gabpα. The expression of several mitochondrial genes that were previously implicated as GABP targets was not significantly affected, but we observed a marked reduction in the expression of the mitochondrial methyltransferase Tfb1m, which is essential for mitochondrial protein translation. We conclude that Gabpα is required for mitochondrial biogenesis and energy production at least in part because of its essential and nonredundant control of Tfb1m expression.
MATERIALS AND METHODS
Cell culture and microscopy.
Mice with loxP recombination sites that flank exons that encode the Gabpa ETS domain (floxed Gabpa [Gabpafl/fl]) were previously described (16). MEFs were prepared from Gabpafl/fl or Gabpa+/+ embryos at embryonic day 12.5 (E12.5) to E14.5 and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (Invitrogen), except where serum starvation is explicitly described. Retroviruses were packaged in the helper-free Phoenix packaging cell line (ATCC, Manassas, VA). For retroviral infection, MEFs at passage 2 were infected with either pBABE-Puro (empty virus) or pBABE-Cre for 48 h and selected for 3 to 5 days in medium containing 2.5 μg/ml puromycin (Sigma-Aldrich, St. Louis, MO). Electron microscopy and fluorescence microscopy were supported by Rhode Island Hospital Core Services.
Semiquantitative RT-PCR and real-time PCR.
Total RNA was prepared with Qiagen (Valencia, CA) RNeasy and reverse transcribed with the Invitrogen Superscript II RT system and poly(dT), and serial 2-fold dilutions of cDNA were subjected to PCR within the linear range of the assay (25 to 35 cycles, depending on the transcript). PCR products were resolved on 1.5% agarose gel, visualized by ethidium bromide, and scanned with an Amersham Typhoon image scanner (Amersham, Piscataway, NJ). Real-time PCR was performed with a Qiagen real-time PCR master kit on a real-time thermal cycler from Stratagene. The sequences of the primers used for reverse transcription (RT)-PCR, real-time PCR, or genomic PCR will be provided upon request.
Analysis of apoptosis.
The Apo-ONE homogeneous caspase 3/7 assay was performed according to the manufacturer's protocol (Promega Corporation, Madison, WI). Staurosporine (Sigma-Aldrich) was applied to cells at 1 μM for 24 to 48 h to induce apoptosis. Intracellular ATP detection was performed according to the manufacturer's protocol (Roche Applied Science).
Mitochondrial quantification and membrane potential.
Mitochondria in wild-type and Gabpα-null MEFs were counted in three separate fields. Mitochondrial mass was evaluated by staining cells with 50 nM NAO (nonyl acridine orange, a membrane potential-insensitive mitochondrial dye; Molecular Probes, Grand Island, NY) or MitoTracker Green FM (Invitrogen) for 30 to 60 min at 37°C (2). Mitochondrial membrane potential was measured by the incubation of MEFs with the MitoTracker probe 5,5′,6,6′-tetrachloro-1,19,3,39-tetraethybenzimidazol carbocyanine iodide (JC-1; Molecular Probes) or MitoTracker Red (Invitrogen) at 5 μg/ml at 37°C for 30 to 60 min (19), followed by flow cytometry or fluorescence microscopy. As a control, wild-type MEFs were incubated with dimethyl sulfoxide or 100 nM valinomycin (a potassium ionophore that dissipates the membrane potential) for 24 h before JC-1 staining.
Western blotting.
Cells were washed in phosphate-buffered saline, and total cellular proteins were prepared by the addition of Laemmli sample buffer, followed by incubation at 100°C for 5 min. Extracts that corresponded to 5 × 105 cells were electrophoresed in 8 to 10% polyacrylamide gel, transferred to Amersham Hybond-P membrane, and probed with antibodies to GABPα, β-actin, procyclic acidic repetitive protein or poly(ADP-ribose) polymerase (PARP), Tomm20, Tomm70 (Santa Cruz Biotechnology), Vdac1 (Cell Signaling), and TFB1M (a gift of Gerald Shadel, Yale University, New Haven, CT).
Oxygen consumption and glycolysis.
Log-phase cells were grown in 24-well XF24 assay plates (Seahorse Bioscience) at 37°C with 5% CO2 for 24 h before analysis. Cells were then incubated with assay medium and transferred to a non-CO2 incubator for 1 h. Oxygen consumption and glycolysis were measured with a Seahorse XF24 analyzer with a Cell Mito Stress Test kit and a glycolysis stress test kit according to the manufacturer's protocols.
ChIP.
Chromatin was immunoprecipitated with antiserum against IgG and GABPα. Chromatin immunoprecipitation (ChIP) was performed as previously described (16).
Pulse-labeling of mitochondrial protein translation.
Labeling of mitochondrial translation in MEFs was performed as previously described (7). Briefly, MEFs were preincubated with methionine-free DMEM for 15 min, followed by methionine-free DMEM for labeling containing 100 μCi/ml 35S (MP Biomedicals, Santa Ana, CA) and 100 μg/ml emetine (Sigma-Aldrich, St. Louis, MO), and chased for 10 min in regular DMEM. Total cellular protein was run on 4 to 20% polyacrylamide gradient gels, which were dried for 1 h and autoradiographed.
RESULTS
Disruption of Gabpα in mice and MEFs.
We used homologous recombination to generate mice in which loxP recombination sites flank exons that encode the Gabpa ets DNA-binding domain (Fig. 1A). Heterozygous floxed (Gabpafl/+) mice were bred with mice that bear the cytomegalovirus-Cre transgene and express Cre recombinase in all of their tissues. The resultant Gabpa+/− mice were phenotypically normal, but intercrossing of these hemizygous mice generated no nullizygous mice among 35 pups. This finding is consistent with previous reports that Gabpα nullizygosity causes an embryonic lethal defect (16, 17).
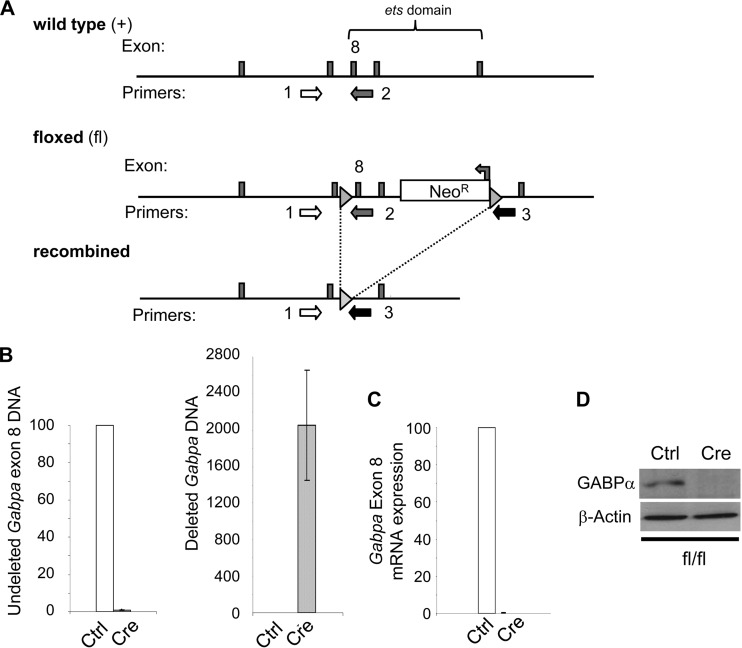
FIG 1.
Disruption of Gabpa in MEFs. (A) Targeting strategy illustrating the wild-type, floxed, and Cre-mediated recombined Gabpa alleles. Exon 8, which encodes part of the Gabpa ETS DNA-binding domain, is indicated; triangles represent loxP sites; arrows indicate PCR primers used for genotyping. (B to D) Analysis of Gabpafl/fl MEFs infected with control (Ctrl) or Cre-expressing (Cre) retrovirus for the presence in genomic DNA of Gabpa exon 8 (with primers 1 and 2 in panel A) and deletion of exon 8 (with primers 1 and 3) by real-time PCR (B), quantitative PCR of reverse-transcribed Gabpa mRNA (C), and immunoblotting to detect Gabpα and β-actin protein expression (D).
We prepared MEFs from wild-type (Gabpa+/+) or homozygous floxed (Gabpafl/fl) embryos and infected them with either pBABE-Cre, which expresses Cre recombinase, or the empty control retrovirus pBABE-Puro. Cre efficiently deleted Gabpa (Fig. 1B), the Gabpa transcript was absent (Fig. 1C), and Gabpα protein was not detected (Fig. 1D) in Gabpafl/fl MEFs (referred to here as Gabpa−/− or Gabpα-null cells).
Gabpα−/− MEFs have a reduced mitochondrial cell mass.
We sought to determine if deletion of Gabpa affected the morphology or the total cellular content of mitochondria. Electron microscopy demonstrated no discernible abnormalities in mitochondrial morphology (Fig. 2A), but there was a statistically significant reduction of mitochondrial content in Gabpα-null MEFs (Gabpafl/fl MEFs infected with pBABE-Cre) compared with that of control cells (Gabpafl/fl MEFs infected with pBABE-Puro) (Fig. 2B) (P < 0.02).
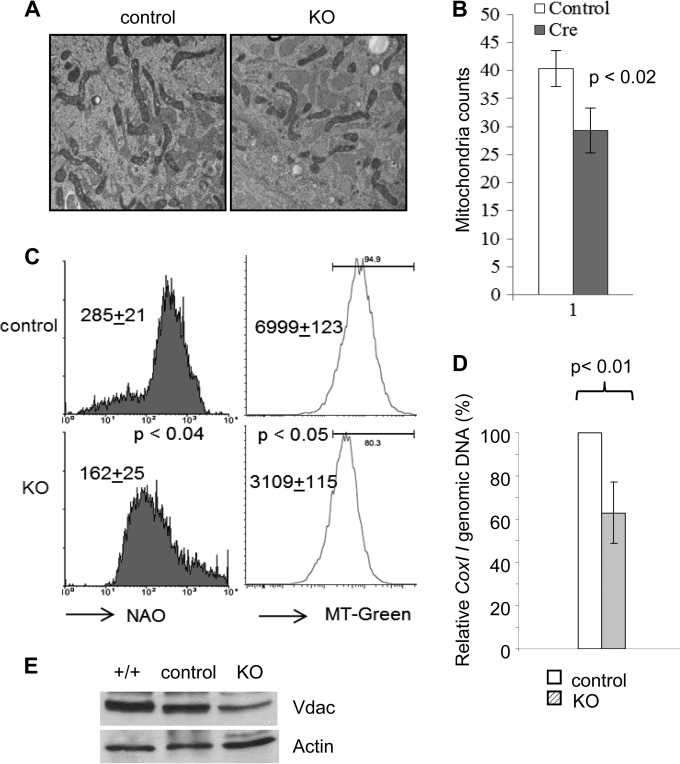
FIG 2.
Gabpa deletion reduces mitochondrial mass in MEFs. (A) Gabpafl/fl MEFs infected with control or Cre-expressing (KO) retrovirus were examined by electron microscopy (×14,000 magnification). (B) Summary of mitochondrial counts per microscopy view (n = 3). (C) Flow cytometry analysis of MEFs stained with NAO (left) and MitoTracker Green (MT-Green, right). Geometric means and P values of the differences are indicated. (D) Ratio of COX I (mitochondrial) to actin B (nuclear) DNA by quantitative real-time PCR. The P values of the differences are indicated. (E) Immunoblotting of retrovirus-infected MEFs with antibodies against Vdac and β-actin.
In order to validate the degree of mitochondrial loss in Gabpα-null MEFs, we incubated cells with NAO, which binds cardiolipin and is retained in mitochondria without regard to their energetic state or membrane potential. NAO staining demonstrated a 40% reduction in mitochondrial mass in Gabpα-null cells, compared to that in control MEFs (Fig. 2C, left) (P < 0.04). We further confirmed this result by staining cells with MitoTracker Green FM, a fluorescent dye that stains live cells and localizes to mitochondria regardless of their membrane potential (Fig. 2C, right) (P < 0.05). As an independent technique for determining the degree of mitochondrial loss, we measured the ratio of mtDNA genes to nuclear genes. The ratio of Cox I (an mtDNA gene) to the gene for actin B (a nuclear gene) was reduced by more than one-third in Gabpα-null cells (Fig. 2D) (P < 0.01). As another indicator of mitochondrial mass, we measured the levels of Vdac1, a gene that encodes a voltage-dependent anion channel protein that is a major component of the mitochondrial outer membrane. Immunoblotting of protein extract from control and knockout (KO) MEFs with antibodies against Vdac1 and β-actin demonstrated a reduction of Vdac1 protein in KO MEFs (Fig. 2E). Thus, these five independent methods demonstrate that Gabpa deletion reduces the cellular content of mitochondria by approximately one-third.
Mitochondrial membrane potential is not altered in Gabpα−/− MEFs.
We stained cells with the cationic dye JC-1 to determine if Gabpα loss impairs the mitochondrial membrane potential. Emission of both orange and green fluorescence by flow cytometry following JC-1 staining indicates mitochondrial membrane potential integrity. Treatment of wild-type MEFs with the antibiotic valinomycin served as a positive control for mitochondrial membrane potential disruption (Fig. 3A). However, we observed no loss of mitochondrial membrane potential in Gabpα-null MEFs, compared to control MEFs (Fig. 3A). As a complementary method, we stained MEFs with MitoTracker Red FM, a fluorescent dye that stains live cells dependent upon their mitochondrial membrane potential. Similar to the result of JC-1 staining, there was no decrease in MitoTracker Red-stained KO MEFs, compared to the control MEFs (Fig. 3B). Mitochondrial membrane potential integrity was confirmed by fluorescence microscopy following JC-1 or MitoTracker Red staining (data not shown). Thus, Gabpa disruption does not impair mitochondrial membrane potential.
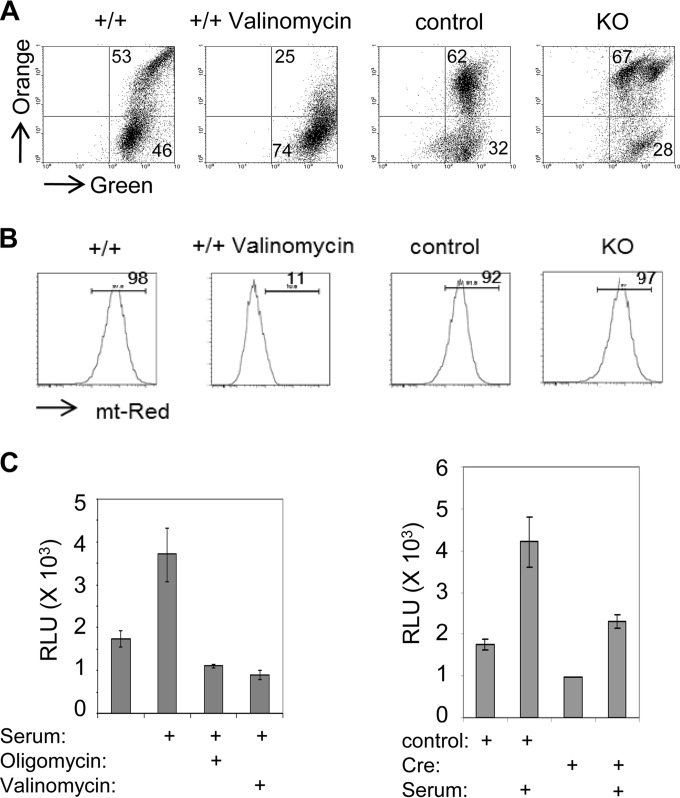
FIG 3.
Mitochondrial function in Gabpa−/− MEFs. Wild-type or Gabpafl/fl MEFs were infected with control or Cre-expressing (KO) retrovirus, stained with MitoTracker JC-1 (A) or MitoTracker Red (mt-Red) (B), and then subjected to flow cytometry to evaluate mitochondrial membrane potential. Valinomycin was used to dissipate the membrane potential and served as a negative control. Values are the percentages of cells in the indicated quadrants or gates. (C) ATP generation measured by luciferase relative light units (RLU) by wild-type MEFs treated with oligomycin or valinomycin (left) or Gabpafl/fl MEFs infected with the control or Cre retrovirus in the absence or presence of 10% fetal calf serum (right).
Reduced energy production in Gabpα−/− MEFs.
Generation of ATP by oxidative phosphorylation and electron transport is the major source of energy generation in the eukaryotic cell. We sought to determine if Gabpa disruption significantly affects cellular energy production. We measured ATP production by luciferase-associated fluorescence, which requires intracellular ATP as an energy source. As a positive control, addition of 10% serum to the growth medium of serum-starved, wild-type MEFs more than doubled ATP production (Fig. 3C, left panel). Treatment with oligomycin (an inhibitor of ATP synthase that is required for oxidative phosphorylation of ADP) or valinomycin abrogated the serum-induced increase in luciferase activity.
We examined ATP production by Gabpα-null MEFs and control cells under conditions of serum starvation and following serum treatment (Fig. 3C, right panel). ATP production by control Gabpafl/fl MEFs infected with pBABE-Puro more than doubled following the addition of serum. Gabpα MEFs exhibited 30 to 40% less ATP production than control MEFs under both serum-starved and serum-treated conditions. Thus, Gabpα−/− MEFs exhibit reduced ATP production at levels that are commensurate with the reduction in mitochondrial mass.
Reduced oxygen consumption in Gabpα−/− MEFs.
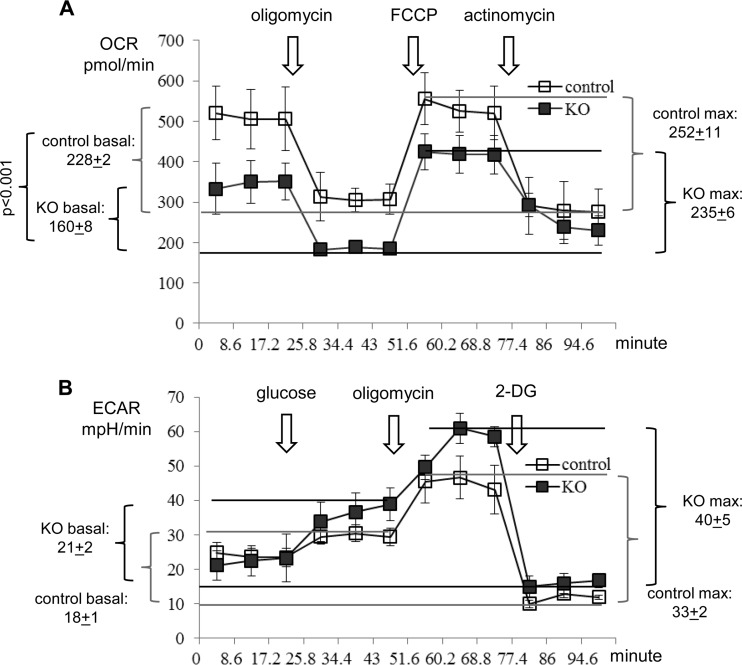
Because cellular ATP is derived from both mitochondrial oxidative phosphorylation and cytoplasmic glycolysis, reduced ATP production in KO MEFs could potentially be due to altered glycolysis. We evaluated both mitochondrial oxygen consumption and the cytoplasmic glycolysis of control and KO MEFs. As shown in Fig. 4A, there was a 30% reduction in the basal oxygen consumption rate (OCR) of KO MEFs (P < 0.001), but the maximal OCRs of control and KO MEFs were comparable. Meanwhile, control and KO MEFs demonstrated similar glycolysis capacities under either resting (basal) or stressed (maximal) conditions (Fig. 4B). Thus, the reduced ATP production was clearly due to decreased oxidative phosphorylation in the mitochondria of Gabpα−/− cells.
FIG 4.
Oxygen consumption and glycolysis in Gabpa−/− MEFs. Shown are the oxygen consumption rate (OCR) (A) and glucose metabolism (B) of Gabpafl/fl (fl/fl) MEFs infected with control or Cre (KO) retrovirus, as measured with a Seahorse XF24 analyzer. ECAR, extracellular acidification rate; FCCP, carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone, the ion-uncoupling reagent; 2-DG, 2-deoxy-d-glucose, an unmetabolizable glucose analog.
No increase in apoptosis in Gabpα−/− MEFs.
Mitochondria are key effectors of programmed cell death through caspase-stimulated release of cytochrome c. Gabpa deletion was reported to increase apoptosis in hematopoietic stem cells (20), but we previously observed no significant increase in apoptosis in Gabpα−/− MEFs, as measured by annexin V and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assays (21). We measured caspase 3/7 activity to further assess apoptosis in Gabpα-null cells. As positive controls, wild-type MEFs treated with staurosporine activated caspase 3/7 (Fig. 5A). However, Gabpα−/− MEFs did not exhibit greater activation of caspase 3/7 than control MEFs.
FIG 5.
Apoptosis following Gabpa deletion. (A) Caspase 3/7 activity of untreated or staurosporine (Stauro)-treated wild-type (+/+) MEFs or Gabpafl/fl (fl/fl) MEFs infected with the control (Ctrl) or Cre retrovirus. (B) Immunoblotting for PARP or β-actin protein from wild-type (+/+) MEFs treated with staurosporine for the indicated numbers of hours and Gabpafl/fl MEFs infected with the control or Cre retrovirus.
We measured caspase-mediated PARP cleavage to further assess apoptosis. As expected, staurosporine treatment of wild-type MEFs increased the level of the 89-kDa cleaved form of PARP, relative to that of 113-kDa PARP (Fig. 5B). However, there was no increase in PARP cleavage in Gabpα-null MEFs, compared to that in controls. We conclude that Gabpa deletion does not increase apoptosis under normal glucose growth conditions.
Mitochondrial gene expression in Gabpα−/− MEFs.
GABP has since been reported to transcriptionally activate or bind the promoters of more than a dozen mitochondrial genes, including the cytochrome c oxidase (COX), F1 ATP synthase β, ATP synthase coupling factor 6 (CF6), and Tfam genes (2). We performed quantitative RT-PCR to assess the expression of putative GABP mitochondrial target genes following Gabpa deletion. Among the components of electron transport and oxidative phosphorylation that we examined (Fig. 6A), the cytochrome c oxidase subunit IV isoform ii (COX IVii; P < 0.005) and COX Vb (P < 0.03) mRNA levels were significantly reduced and the COXIVi mRNA level was increased (P < 0.03) following Gabpa disruption, but there was no significant alteration of F1 ATP synthase β, ATP synthase CF6, or cytochrome c mRNA levels compared to those of control MEFs.
FIG 6.
Expression of key mitochondrial genes and proteins in Gabpa−/− MEFs. Gabpafl/fl MEFs infected with the control (Ctrl) or Cre retrovirus were analyzed by quantitative RT-PCR of key mitochondrial genes (A) and transcription factors and cofactors (B) that affect mitochondrial biogenesis. Cyto C, cytochrome c; *, P < 0.03; **, P < 0.01. (C) Analysis of protein expression from these MEFs by immunoblotting for Tfb1m and β-actin. (D) ChIP of F1 ATP synthase β and ATP synthase cofactor 6 with antibodies against IgG, GABPα (α), or GABPβ (β). neg, no DNA; input, genomic DNA. (E) Wild-type and Gabpafl/fl MEFs infected with the control or Cre retrovirus analyzed by immunoblotting for the expression of mitochondrial membrane proteins Tomm20 and Tomm70. (F) Analysis of mitochondrial protein synthesis in these MEFs by radioactive 35S pulse-labeling in the presence of emetine. Immunoblotting for β-actin indicated equal loading of total cellular proteins. Mitochondrial (mt) translation products are identified on the left.
We examined the expression of the mRNAs of transcription factors and cofactors that are known to regulate mitochondrial gene expression or biogenesis. There was no significant change in the Tfam, Tfb2m, Nrf1, c-Myc, Pgc-1a, or Prc mRNA level (Fig. 6B). However, the expression of Tfb1m mRNA was decreased by more than 70% in Gabpα-null cells (P < 0.005). The reduction of Tfb1m protein in Gabpα-null MEFs was confirmed by immunoblotting (Fig. 6C). Both Gabpα and Gabpβ bound to the promoters of F1 ATPase β and ATPase CF6 in control cells, but neither component of the GABP complex bound in Gabpα-null cells (Fig. 6D); this is consistent with our previous observation that GABPβ cannot bind to ETS sites in the Cox Vb promoter in the absence of GABPα (18). We conclude that only some of its putative mitochondrial gene targets are affected by Gabpa disruption but that the ribosomal methyltransferase Tfb1m is dependent on Gabp for expression.
TFB1M is a limiting factor that controls mitochondrial protein synthesis (7, 8), and reduced Tfb1m expression after Gabpa deletion might account for the decreased mitochondrial biogenesis in Gabpα-null MEFs. We examined total mitochondrial protein synthesis by 35S pulse-labeling and observed decreased synthesis of all mitochondrial proteins in Gabpα-null MEFs; the reduction in mitochondrial proteins was global and not limited to any specific mitochondrial protein (Fig. 6E). We also examined the possibility that protein transport defects account for reduced mitochondrial biogenesis by examining the mitochondrial membrane proteins Tomm20 and Tomm70, which are crucial for mitochondrial protein transport across the mitochondrial membrane. Neither Tomm20 nor Tomm70 protein expression was affected by Gabpa deletion, as shown in Fig. 6F. Together with the previous report that decreased Tfb1m resulted in decreased mitochondrial protein translation and mitochondrial mass (7), we conclude that Gabp regulates mitochondrial biogenesis through its control of the key mitochondrial factor Tfb1m.
DISCUSSION
Coordination of the transcription and translation of electron transport chain genes is challenging for the cell because various components are located in either the nuclear or the mitochondrial genome (2). GABP is an ETS-related tetrameric transcription factor (also known as nuclear respiratory factor 2 or NRF2) (11) that has been implicated as a regulator of genes involved in the respiratory chain and oxidative phosphorylation. GABP is one of more than two dozen mammalian ETS factors that recognize and bind similar cognate DNA sequences (12), but to date, it has not been clear if GABP plays an essential, nonredundant role in the regulation of mitochondrial genes.
We genetically disrupted Gabpa and examined its role in the control of mitochondrial function, biogenesis, and gene expression in MEFs. Gabpα loss did not disrupt mitochondrial structure, alter membrane potential, or increase apoptosis. However, mitochondrial mass was reduced by one-third in Gabpα-null MEFs, and there was a commensurate decrease in ATP production, oxygen consumption, and overall mitochondrial protein synthesis. Among the genes involved in electron transport and oxidative phosphorylation, only COX Vb and Cox IVii (a minor isoform that is expressed primarily in lung tissue) were affected by Gabpa disruption. Several transcription factors and cofactors have been shown to regulate mitochondrial biogenesis and gene expression. Expression of the transcription factor Tfb1m decreased significantly following Gabpα loss, and overall mitochondrial protein synthesis decreased commensurately. We observed a more restricted set of biologically significant mitochondrial target genes than has been previously proposed (2), but methodological (our use of gene disruption rather than an RNA interference) or species differences may explain the disparate results. We conclude that GABP has a critical, nonredundant role in the regulation of mitochondrial biogenesis and energy production and that it plays an essential role in the control the expression of Tfb1m, which itself is required for mitochondrial biogenesis.
The 16-kb mitochondrial genome does not encode any of the transcription factors and cofactors that are needed for mtDNA gene transcription. Indeed, the mitochondrial genome contains 37 genes and encodes only 13 proteins of the >80 protein subunits that are necessary for electron transport and oxidative phosphorylation. Thus, mtDNA gene expression depends on nuclear transcription factors, including GABP. Loss of mouse Gabpα caused early, peri-implantation embryonic lethality (16–18). Gabp is both necessary and sufficient for cell cycle entry (16), and genetic disruption of Gabpa in mouse bone marrow caused a profound loss of hematopoietic progenitor cells (20, 22) and defects in white blood cell development (16). Thus, GABP plays essential roles in the cell cycle, proliferation, and differentiation that cannot be replaced by other ETS factors. We now demonstrate that Gabp is required for mitochondrial biogenesis and energy production and that it plays a nonredundant role in the control of Tfb1m expression.
Mitochondrial gene transcription requires a unique RNA polymerase (POLRMT), but it is capable of only modest transcriptional activity without TFAM and TFBM (1, 4). POLRMT is the sole RNA polymerase in mitochondria, but it is capable of only modest transcriptional activity without TFB2M and TFAM. Genetic disruption of Tfam caused severe respiratory chain deficiency, mtDNA depletion, large aberrant mitochondria with poorly defined cristae, and early embryonic lethality (23). Similarly, disruption of the transcription termination factor mTERF3 or deletion of PolgA, a subunit of mtDNA polymerase, caused profound mitochondrial defects and embryonic lethality at E8.5 (24, 25). Thus, early embryonic lethality and severe respiratory chain deficiencies resulted from loss of nuclear proteins that function exclusively in mitochondrial gene expression or mtDNA maintenance.
Disruption of transcription factors whose activities are not restricted to mitochondria caused distinctly different mitochondrial phenotypes. Genetic disruption of NRF-1 caused mtDNA depletion and loss of mitochondrial membrane potential, but nonmitochondrial defects appeared to be the cause of early embryonic lethality (26). The YY1 initiator element binding factor controls certain cytochrome oxidase subunit genes either positively or negatively (4, 27), and its disruption caused early embryonic lethality (28). c-Myc is a key regulator of the cell cycle and metabolic networks (29), and it stimulates the expression of certain NRF1 target genes (10). Embryos that lack c-Myc are lost between E9.5 and E10.5 (30), and c-myc-null fibroblasts are deficient in mitochondria (31). Disruption of other factors implicated in mitochondrial biogenesis, including PPARα, the orphan nuclear receptor ERRα, and the coactivators PGC-1a and PGC1b caused more subtle mitochondrial defects (21, 32–38). Thus, transcriptional regulators that control both mitochondrial and nuclear genes appear to modulate, rather than fundamentally control, mitochondrial biogenesis and function.
TFB1M and TFB2M were previously thought to be mitochondrial transcription factors (6, 19), but in vivo and in vitro studies have revised our understanding of their functions and activities (5, 6). Both TFB1M and TFB2M are mammalian RNA dimethyltransferases that modify conserved adenines in the mitochondrial small ribosomal subunit rRNA. Both TFB1M and TFB2M play crucial, nonredundant roles in the control of mitochondrial biogenesis. Overexpression of TFB2M significantly increased mtDNA and mitochondrial gene transcription, while reduced levels of TFB2M decreased the quantity of mtDNA and reduced mitochondrial gene transcription (7, 9). Although overexpression of TFB1M did not affect mitochondria (7, 8), reduced levels of TFB1M significantly decreased overall mitochondrial protein translation because of insufficient rRNA methylation (7, 9). Thus, TFB1M is limiting for mitochondrial biogenesis and the phenotype associated with reduced TFB1M resembles the mitochondrial defects caused by Gabpα disruption.
Gabpa disruption did not cause the profound mitochondrial phenotype that is associated with loss of factors that function solely in mitochondria, such as Tfam, TERF3, and PolgA. Furthermore, Gabpa disruption caused very early embryonic lethality (39), which suggests that embryonic loss was not caused solely by its role in mitochondrial regulation. Gabpa disruption dramatically reduced Tfb1m expression, and the phenotype of reduced mitochondrial biogenesis was similar to that caused by short hairpin RNA knockdown of TFB1M. Because reduced expression of TFB1M phenocopies Gabpa deletion, this suggests that GABP regulates mitochondrial biogenesis mainly through its transcriptional control of TFB1M. Curiously, among the mitochondrial genes that we examined, Gabpa disruption reduced the expression of only CoxIVii and CoxVb. It is unclear if there is something distinctive about the promoter or other regulatory elements of these two genes that does not permit other transcription factors to compensate for the loss of Gabp, as must occur with the other mitochondrial target genes whose expression is not significantly affected by Gabpα loss. In summary, we conclude that GABP is a key regulator of both nuclear and mitochondrial genes and plays an essential, nonredundant role in mitochondrial biogenesis through its control of the RNA dimethyltransferase TFB1M.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 HL073945 to A.G.R.
We thank Gerald Shadel (Yale University) for TFB1M antibody and Marcus Cooper (University of Massachusetts) for use of the Seahorse XF24 analyzer.
Footnotes
Published ahead of print 23 June 2014
REFERENCES
- 1.Bonawitz ND, Clayton DA, Shadel GS. 2006. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell 24:813–825. 10.1016/j.molcel.2006.11.024 [DOI] [PubMed] [Google Scholar]
- 2.Ongwijitwat S, Wong-Riley MT. 2005. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene 360:65–77. 10.1016/j.gene.2005.06.015 [DOI] [PubMed] [Google Scholar]
- 3.Scarpulla RC. 2008. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N. Y. Acad. Sci. 1147:321–334. 10.1196/annals.1427.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scarpulla RC. 2011. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta 1813:1269–1278. 10.1016/j.bbamcr.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. 2010. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285:18129–18133. 10.1074/jbc.C110.128918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCulloch V, Seidel-Rogol BL, Shadel GS. 2002. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 22:1116–1125. 10.1128/MCB.22.4.1116-1125.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotney J, McKay SE, Shadel GS. 2009. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet. 18:2670–2682. 10.1093/hmg/ddp208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotney J, Wang Z, Shadel GS. 2007. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 35:4042–4054. 10.1093/nar/gkm424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsushima Y, Adán C, Garesse R, Kaguni LS. 2005. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J. Biol. Chem. 280:16815–16820. 10.1074/jbc.M500569200 [DOI] [PubMed] [Google Scholar]
- 10.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. 2009. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9:386–397. 10.1016/j.cmet.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 11.Virbasius JV, Virbasius CA, Scarpulla RC. 1993. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 7:380–392. 10.1101/gad.7.3.380 [DOI] [PubMed] [Google Scholar]
- 12.Hsu T, Trojanowska M, Watson DK. 2004. Ets proteins in biological control and cancer. J. Cell. Biochem. 91:896–903. 10.1002/jcb.20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. 2004. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol. Dis. 32:143–154. 10.1016/j.bcmd.2003.09.005 [DOI] [PubMed] [Google Scholar]
- 14.Luo M, Shang J, Simkevich CP, Jackson CL, King TC, Rosmarin AG. 1999. Characterization and localization to chromosome 7 of psihGABPalpha, a human processed pseudogene related to the ets transcription factor, hGABPalpha. Gene 234:119–126. 10.1016/S0378-1119(99)00167-5 [DOI] [PubMed] [Google Scholar]
- 15.Brown TA, McKnight SL. 1992. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 6:2502–2512. 10.1101/gad.6.12b.2502 [DOI] [PubMed] [Google Scholar]
- 16.Yang Z-F, Mott S, Rosmarin AG. 2007. The Ets transcription factor GABP is required for cell-cycle progression. Nat. Cell Biol. 9:339–346. 10.1038/ncb1548 [DOI] [PubMed] [Google Scholar]
- 17.Ristevski S, O'Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. 2004. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol. Cell. Biol. 24:5844–5849. 10.1128/MCB.24.13.5844-5849.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, Williams A, McCoy JP, Leonard WJ. 2004. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat. Immunol. 5:1036–1044. 10.1038/ni1117 [DOI] [PubMed] [Google Scholar]
- 19.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson 2002. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31:289–294. 10.1038/ng909 [DOI] [PubMed] [Google Scholar]
- 20.Yang ZF, Drumea K, Cormier J, Wang J, Zhu X, Rosmarin AG. 2011. GABP transcription factor is required for myeloid differentiation, in part, through its control of Gfi-1 expression. Blood 118:2243–2253. 10.1182/blood-2010-07-298802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, Shulman GI, Spiegelman BM, Lowell BB. 2006. Hypomorphic mutation of PGC-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 4:453–464. 10.1016/j.cmet.2006.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu S, Cui K, Jothi R, Zhao DM, Jing X, Zhao K, Xue HH. 2011. GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood 117:2166–2178. 10.1182/blood-2010-09-306563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231–236 [DOI] [PubMed] [Google Scholar]
- 24.Hance N, Ekstrand MI, Trifunovic A. 2005. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 14:1775–1783. 10.1093/hmg/ddi184 [DOI] [PubMed] [Google Scholar]
- 25.Park CB, Asin-Cayuela J, Cámara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG. 2007. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell 130:273–285. 10.1016/j.cell.2007.05.046 [DOI] [PubMed] [Google Scholar]
- 26.Huo L, Scarpulla RC. 2001. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol. Cell. Biol. 21:644–654. 10.1128/MCB.21.2.644-654.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A, Lenka N, Mullick J, Avadhani NG. 1997. Regulation of murine cytochrome oxidase Vb gene expression in different tissues and during myogenesis. Role of a YY-1 factor-binding negative enhancer. J. Biol. Chem. 272:5899–5908 [DOI] [PubMed] [Google Scholar]
- 28.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19:7237–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. 2008. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7:1054–1066. 10.4161/cc.7.8.5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671–682 [DOI] [PubMed] [Google Scholar]
- 31.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25:6225–6234. 10.1128/MCB.25.14.6225-6234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. 2005. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 1:259–271. 10.1016/j.cmet.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 33.Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM. 2007. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J. Clin. Invest. 117:3463–3474. 10.1172/JCI31785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huss JM, Torra IP, Staels B, Giguère V, Kelly DP. 2004. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24:9079–9091. 10.1128/MCB.24.20.9079-9091.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. 2008. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 22:1948–1961. 10.1101/gad.1661708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15:3012–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly-Y M, Storlien L, Strömstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A. 2006. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 4(11):e369. 10.1371/journal.pbio.0040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. 2005. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3(4):e101. 10.1371/journal.pbio.0030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian Y, Banerjee S, Grossman CE, Amidon W, Nagy G, Barcza M, Niland B, Karp DR, Middleton FA, Banki K, Perl A. 2008. Transaldolase deficiency influences the pentose phosphate pathway, mitochondrial homoeostasis and apoptosis signal processing. Biochem. J. 415:123–134. 10.1042/BJ20080722 [DOI] [PubMed] [Google Scholar]