Significance
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 nuclease (Cas9) system has recently emerged as an efficient and versatile tool for genome editing in various organisms. However, its targeting capability and multiplex editing efficiency are often limited by the guide RNA (gRNA)-expressing device. This study demonstrates a general strategy and platform for precise processing and efficient production of numerous gRNAs in vivo from a synthetic polycistronic gene via the endogenous tRNA-processing system. This strategy is shown to significantly increase CRISPR/Cas9 multiplex editing capability and efficiency in plants and is expected to have broad applications for small RNA expression and genome engineering.
Keywords: CRISPR/Cas9, tRNA processing, genome editing, multiplex
Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 nuclease (Cas9) system is being harnessed as a powerful tool for genome engineering in basic research, molecular therapy, and crop improvement. This system uses a small guide RNA (gRNA) to direct Cas9 endonuclease to a specific DNA site; thus, its targeting capability is largely constrained by the gRNA-expressing device. In this study, we developed a general strategy to produce numerous gRNAs from a single polycistronic gene. The endogenous tRNA-processing system, which precisely cleaves both ends of the tRNA precursor, was engineered as a simple and robust platform to boost the targeting and multiplex editing capability of the CRISPR/Cas9 system. We demonstrated that synthetic genes with tandemly arrayed tRNA–gRNA architecture were efficiently and precisely processed into gRNAs with desired 5′ targeting sequences in vivo, which directed Cas9 to edit multiple chromosomal targets. Using this strategy, multiplex genome editing and chromosomal-fragment deletion were readily achieved in stable transgenic rice plants with a high efficiency (up to 100%). Because tRNA and its processing system are virtually conserved in all living organisms, this method could be broadly used to boost the targeting capability and editing efficiency of CRISPR/Cas9 toolkits.
Higher organisms often use complicated genetic networks with functionally redundant genes to fine tune cellular processes. Therefore, molecular tools with the capability to simultaneously manipulate multiple genes are of great value in both basic research and practical applications of genetic engineering. Recently, the bacterial type II clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (Cas) protein system is emerging as a promising tool for this purpose. The Cas9 endonuclease from Streptococcus pyogenes, coupled with an artificial guide RNA (gRNA), is able to target the DNA sequence of 5′-N20-NGG-3′ (N indicates any base), in which N20 is the same as 20 bases of the gRNA 5′ sequence (referred to as gRNA spacer hereafter) and NGG is the protospacer-adjacent motif (PAM) (1–3). The simple RNA-guided programmable rule and high occurrence of PAM in genomes allowed Cas9-gRNA to readily target almost all genetic elements for genome editing. Owing to its simplicity and high efficiency, Cas9-based tools have been rapidly developed for genome/epigenome editing, transcriptional regulation, and other applications in genetic engineering (4–6).
In principle, multiplex genome editing could be achieved by expressing Cas9 (or Cas9-derived effectors) along with multiple gRNAs for respective target sites. Conventional delivery methods involve either microinjection or expression of gene constructs containing multiple single gRNA (sgRNA)-expressing cassettes for multiplex genome editing (2, 3, 7–11). Direct microinjection of in vitro-synthesized gRNAs and Cas9 protein (or Cas9 mRNA) into the cell or embryo is suitable for only very few systems. As a result, the most common strategy is to stack multiple sgRNA-expressing cassettes in one plasmid construct or to use multiple constructs. The typical size of one sgRNA-expressing cassette is about 400–500 bp and consists of the RNA polymerase III (Pol III) promoter, gRNA, and the Pol III terminator. Due to the limitation of delivery method and plasmid vector capacity, simultaneous production of numerous gRNAs would be a challenge with such a gRNA-expressing strategy for most organisms. Moreover, eukaryotic Pol III-transcribed RNA is obligated to start with a specific ribonucleotide, which may reduce the Cas9/gRNA targetable sites. An advanced strategy would be to compact multiple gRNAs in one synthetic gene and engineer an RNA-processing system to excise individual gRNAs from the primary transcript. The only proven method based on such a strategy is the expression of Csy4 endoribonuclease (endo-RNase) with Cas9 to process a transcript containing gRNAs fused with Csy4-cleavable RNA (12, 13). Nevertheless, more robust and precise methods for simultaneous production of multiple gRNAs are needed to improve multiplex editing capability and facilitate more sophisticated Cas9 applications.
RNA is a fundamental cellular component, and its production is guaranteed by the conserved and precise RNA-processing systems in different organisms. This notion inspired us to engineer an endogenous RNA-processing system to produce multiple gRNAs from a single transcript, rather than introducing any additional RNases along with Cas9/gRNA components. In this study, we demonstrated that multiple gRNAs could be efficiently produced from a single synthetic gene with the tRNA–gRNA architecture after precise excision of transcripts in vivo by the endogenous RNases. This gRNA-expressing strategy was shown to not only allow multiplex targeting but also improve the editing efficiency of the CRISPR/Cas9 system in plants. Because the tRNA-processing system exists in virtually all organisms, this strategy could be broadly used to boost the multiplex editing capability of CRISPR/Cas9 tools for genome engineering.
Results
Strategy to Engineer a tRNA-processing System for Producing Numerous gRNAs.
To simultaneously produce multiple gRNAs from one primary transcript, we aimed to compact a cluster of gRNAs with different spacers in one polycistronic gene and hijack an endogenous RNA-processing system for cleavage of this transcript into individual gRNAs in a nucleus. In search of RNA and endo-RNases that potentially meet this requirement, tRNA attracted our attentions for its characteristics as follows. (i) In nucleoplasm, the tRNA precursors (pre-tRNAs) are cleaved at specific sites in eukaryotes by RNase P and RNase Z (or RNase E in bacterium) to remove 5′ and 3′ extra sequences (Fig. 1A), respectively (14–18). (ii) RNase P and RNase Z recognize the tRNA structure, regardless of the pre-tRNA sequence (17, 18). Previous studies revealed that only the acceptor stem, D-loop arm, and TψC-loop arm of tRNA are necessary for RNase P and RNase Z cleavage of pre-tRNA (16–19). (iii) tRNA was found in the polycistronic transcription unit in bacteria (20) and occasionally in eukaryotes (19, 21), suggesting that the tRNA-processing system is likely used as an intrinsic mechanism to produce different small RNAs [e.g., small nucleolar RNA (snoRNA)] with tRNA from a single polycistronic gene (19). (iv) Because tRNA is one of the most abundant cellular components, the endogenous tRNA-processing system should be robust to process a large number of substrates. (v) The tRNA genes contain internal promoter elements (box A and B) to recruit the RNA polymerase III (Pol III) complex (Fig. 1B) (22, 23). Therefore, the tRNA sequence may also work as a potential transcriptional enhancer for Pol III. Based on these features, we hypothesized that the endogenous tRNA system could be engineered as a general platform for precise processing of gRNAs.
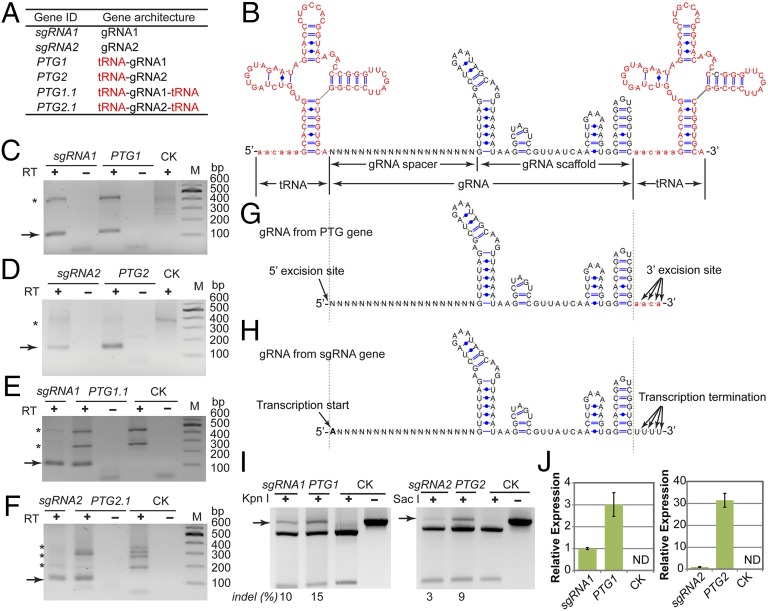
Fig. 1.
Engineering the endogenous tRNA system for multiplex genome editing with CRISPR/Cas9. (A) The eukaryotic pre-tRNA with 5′ leader and 3′ trailer is cleaved by RNase P and RNase Z at specific sites. (B) Transcription of tRNA gene with RNA polymerase III (Pol III). The box A and box B elements in the tRNA gene function as internal transcriptional elements and are bound by transcription factor IIIC (TFIII C), which recruits TFIIIB and Pol III to start transcription. (C) Schematic depiction of the PTG/Cas9 method for simultaneously targeting multiple sites. The synthetic PTG consists of tandemly arrayed tRNA-gRNA units, with each gRNA containing a target-specific spacer (labeled as a diamond with different color) and conserved gRNA scaffold (rectangle). The tRNA containing box A and B elements is shown as round rectangles. The primary transcript of PTG is cleaved by endogenous RNase P and RNase Z (labeled as scissors) to release mature gRNAs and tRNA (red lines of cloverleaf structure). The excised mature gRNAs direct Cas9 to multiple targets.
To use the endogenous tRNA system for multiplex genome editing with Cas9/gRNA, we designed a polycistronic tRNA-gRNA (PTG) gene for simultaneous production of numerous gRNAs. As shown in Fig. 1C, this PTG gene consists of tandem repeats of tRNA-gRNA and would be transcribed as a normal snoRNA or sgRNA gene under the control of the Pol III promoter. The endogenous tRNA-processing RNases (e.g., RNase P and RNase Z in plants) would recognize the tRNA components and excise individual gRNAs from the PTG transcript. The resulting gRNAs would then direct Cas9 to multiple target sites for genome editing (Fig. 1C).
Precise Processing of PTG to Produce Functional gRNAs with Desired Targeting Sequences.
To explore whether the synthetic PTG gene would be transcribed, would be processed, and would function as we predicted (Fig. 1C), we synthesized four PTG genes with a structure of tRNA-gRNA (PTG1 and PTG2) or tRNA-gRNA-tRNA (PTG1.1 and PTG2.1) (Fig. 2A). These four PTG genes were designed to produce gRNA1 (PTG1 and PTG1.1) and gRNA2 (PTG2 and PTG2.1), which have been tested previously (24) with sgRNA genes (sgRNA1 and sgRNA2) in Cas9-mediated genome editing (SI Appendix, Fig. S1A; and see gene sequence in SI Appendix, Table S1). Both gRNAs targeted the rice MPK5 gene, which encodes a mitogen-activated protein kinase (MAPK) involved in biotic and abiotic signaling pathways. These PTGs were constructed with a 77-bp long pre-tRNAGly gene, which recognizes the GGC codon and is widely present among various genomes (25). The chosen pre-tRNAGly sequence consists of a 5′ leader (5′-AACAAA-3′, 6 bp) and mature tRNA (71 bp) (Fig. 2B). Such tRNA–gRNA fusion in PTGs mimics the native tRNA–snoRNA43 dicistron in plants (19).
Fig. 2.
Precise excision of functional gRNAs in vivo from synthetic PTG genes. (A) The architecture of two sgRNA genes and four PTGs to produce one gRNA. (B) Sequence and predicted secondary structure of tRNA–gRNA–tRNA fusion of PTG gene. The bases of the tRNA region are indicated with red color and the tRNA 5′ leader is shown in lowercase. The gRNA is indicated in black, and the gRNA spacer sequence is shown as N. (C–F) Examination of mature gRNAs produced from sgRNA or PTGs with cRT-PCR. Total RNAs from the protoplasts expressing empty vector were used as control (CK). Arrows indicate mature gRNAs amplified by cRT-PCR, and asterisks indicate the nonspecifically amplified rRNA. (G) Summary of excision sites in PTG according to mapped gRNA ends from cRT-PCR (SI Appendix, Figs. S3–S5). Arrows indicate the cleavage sites in PTG to release gRNA. The mature gRNA 5′ ends were excised from PTG exactly at the tRNA–gRNA fusion site in all cRT-PCR results whereas its 3′ ends shifted 1–4 nt within the tRNA 5′ leader (lowercase). (H) gRNA produced from U3p:sgRNA. All detected U3p:sgRNA-produced gRNA started with ribonucleotide A and terminated with multiple Us. (I) Introduction of indels at the desired sites by PTG1:Cas9 or PTG2:Cas9 in rice protoplasts as shown by PCR/RE. Arrows indicate mutated fragments resistant to RE digestion. The indel frequency is indicated at the bottom. (J) Relative expression of sgRNA1/2 and PTG1/2 in rice protoplasts. Data represent mean ± SD. ND, not detected. CK, empty vector control.
In this study, we used a plasmid vector (SI Appendix, Fig. S2) in which sgRNA or PTG is expressed with the rice U3 snoRNA promoter (U3p) and Cas9 is expressed with a rice ubiquitin promoter plus the complete 5′ untranslated region (UBIp). After transfecting rice protoplasts with plasmids containing U3p:sgRNA or U3p:PTG, circularized reverse transcription PCR (cRT-PCR) (26, 27) was performed to map both 5′ and 3′ ends of mature gRNAs (see SI Appendix, Fig. S3 and Table S2 for the principle of cRT-PCR). Mature gRNAs with the expected size were detected from the protoplasts expressing PTGs or sgRNAs (Fig. 2 C–F). However, the tRNA–gRNA fused transcript from PTGs was not detectable by cRT-PCR, probably due to the highly robust tRNA-processing system that cleaved most (if not all) primary transcripts of PTGs. Sequence analysis of the cRT-PCR products revealed that all four PTG transcripts were precisely cleaved at the tRNA–gRNA junction and produced mature gRNAs carrying the desired 5′ spacer sequences without extra nucleotides (Fig. 2G and SI Appendix, Figs. S4 and S5). On the other hand, the 3′ end of mature gRNAs carried a 1- to 7-nt-long poly (U) tail if it preceded the Pol III terminator (sgRNA, PTG1, and PTG2) (SI Appendix, Fig. S4), or 1- to 4-nt ribonucleotides from the tRNA-leader when it preceded the tRNA (PTG1.1 and PTG2.1) (SI Appendix, Fig. S5). The cRT-PCR data also confirmed that gRNAs transcribed from U3p:sgRNA were obligated to start with nucleotide A whereas U3p:PTGs produced gRNAs without constraint of the first nucleotide (SI Appendix, Figs. S4 and S5). In summary, the gRNAs produced from PTG are the same as the sgRNAs but are not obligated to start with a specific nucleotide (Fig. 2 G and H).
The functionality of PTG1 and PTG2 was confirmed by examining the insertion/deletion (indel) mutations introduced by nonhomologous end joining (NHEJ) repairing at the predicted Cas9:gRNA cleavage site. Because gRNA1 and gRNA2 targets contain the KpnI and SacI restriction enzyme (RE) sites (SI Appendix, Fig. S1A), respectively, the mutations induced by PTG1/Cas9 and PTG2/Cas9 could be readily analyzed by the digestion of PCR products encompassing the targeted sites with corresponding RE (PCR/RE assay). In rice protoplasts transfected with PTG1/Cas9 and PTG2/Cas9, 15% and 9% of the target sites were found to carry indels, respectively (Fig. 2I), which are slightly higher than the mutation rate of the sgRNA:Cas9 constructs we used previously (24). Consistent with our hypothesis that tRNA may function as a transcriptional enhancer for Pol III, the quantitative RT-PCR with gRNA-specific primers revealed that the transcript levels of PTG1 and PTG2 were about 3 and 31 times higher than those of sgRNA1 and sgRNA2 in protoplasts (Fig. 2J), respectively. Taken together, our results demonstrated that the endogenous tRNA system could be used as a precise and robust tool to produce gRNAs from PTG for Cas9-mediated genome editing.
Efficient Multiplex Genome Editing in Rice Protoplasts via PTG/Cas9.
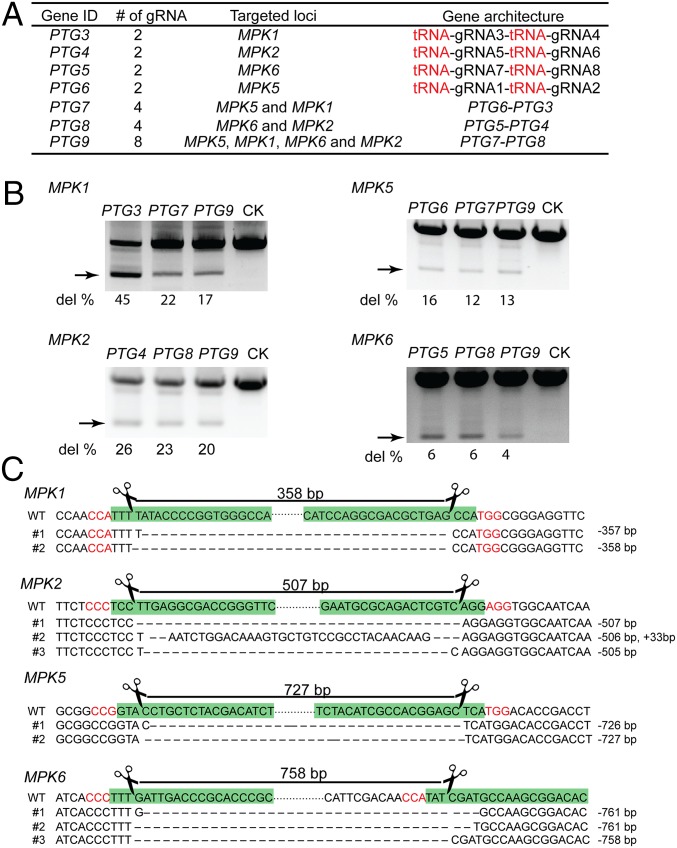
To test the targeting capacity and efficiency of the PTG system, up to eight gRNAs (gRNA1–gRNA8) (SI Appendix, Table S3 and Fig. S1) were tandemly arrayed to construct PTGs for targeting four homologous rice MAPKs (MPK1/2/5/6), which may function redundantly in diverse cellular signaling pathways. These gRNAs were divided into four pairs and each pair targeted two genomic sites within a gene locus with a distance of 350–750 bp between them (SI Appendix, Fig. S1). By combining different gRNA pairs, we designed PTG genes encoding two (PTG3/4/5/6), four (PTG7/8), and eight (PTG9) gRNAs to simultaneously target one, two, and four MPK loci, correspondingly (Fig. 3A and SI Appendix, Table S1). We expected that such a design might result in a deletion of a short chromosomal fragment between two Cas9 cut sites and allow us to readily examine the efficacy of PTGs.
Fig. 3.
Simultaneous editing of multiple genomic sites in rice protoplasts expressing PTG:Cas9. (A) Architecture, gRNA components, and targets of PTGs for multiplex genome editing. (B) PCR detection of chromosomal fragment deletion at targeted loci in rice protoplasts expressing respective PTGs with Cas9. Successful deletion is shown as truncated PCR product (indicated with arrows). The chromosomal fragment deletion frequency (del %) is indicated at the bottom of each lane. The protoplast samples expressing an empty vector were used as control (CK). (C) Representative sequences of chromosomal fragment deletion aligned with that of WT. The gRNA paired region is labeled with green color, and the PAM region is shown in red color letters. The number at the end indicates deleted (−) or inserted (+) bases between two Cas9 cuts. The total length between two Cas9 cut sites (labeled with scissor) is indicated on the top. Short lines in the aligned sequences indicate deletions.
To synthesize PTG with repetitive tRNA—gRNA architecture, we designed a scalable and flexible approach to assemble PTGs from PCR components based on the principle of Golden Gate assembly strategy (28) (see SI Appendix, SI Methods and Figs. S6 and S7 for details). Our gene-assembly approach allowed fast synthesis of PTGs with different combination of gRNAs from common oligonucleotide primers. Using a one- or two-step assembly, we synthesized PTG3-PTG9 genes and cloned them into the CRISPR/Cas9-expressing vectors (SI Appendix, Fig. S2 and Tables S1–S3).
We then transfected rice protoplasts with these plasmids containing both U3p:PTG3-PTG9 and UBIp:Cas9. Chromosomal fragment deletions at four MAPK loci, which were revealed by amplification of truncated PCR products with target specific primers, were detected in protoplasts expressing respective PTGs with 4–45% efficiency (Fig. 3B). Even though eight gRNAs were simultaneously produced from PTG9, they still guided Cas9 to efficiently excise chromosomal fragments at all four targeted loci with 4–20% frequency (Fig. 3B). The fragment deletion efficiency at MPK5 loci was further confirmed by quantitative PCR using primers encompassing the gRNA2 cut site (SI Appendix, Table S4). Sequencing of these truncated PCR products confirmed that fragments between two gRNA/Cas9 cut sites were excised from targeted loci with or without additional indels (Fig. 3C). In general, the fragment deletion efficiency was negatively correlated to the distance between two paired cut sites (correlation coefficient r = −0.95) despite the fact that different gRNAs may have variable efficiencies. We noticed that the total number of gRNAs in one PTG affected the deletion efficiency with variable extent at different targets in protoplasts (Fig. 3B). At three targeting loci (MPK2/5/6), the PTGs (PTG4/5/6) containing two gRNAs exhibited only slightly higher efficiency than the PTGs with four (PTG7/8) and eight (PTG9) gRNAs. However, at the MPK1 locus, PTG3 (two gRNAs) showed ∼2 times higher deletion frequency than PTG7 (four gRNA) or PTG9 (eight gRNAs). Such a reduction of efficiency of PTGs with a higher number of gRNAs was likely due to the competition for Cas9 among gRNAs. Nevertheless, PTG with tandemly arrayed tRNA–gRNA architecture is capable of simultaneously producing numerous gRNAs and guiding Cas9 to multiple chromosomal targets with a high efficiency.
Improving Multiplex Genome Editing in Stable Transgenic Plants with PTG/Cas9.
Because many plants, including important crops like rice, could not be readily regenerated from protoplasts, we used the conventional Agrobacterium-mediated transformation to produce stable transgenic lines and evaluated the efficacy of PTG/Cas9 system for multiplex genome editing in intact rice plants. Mutagenesis frequency at gRNA1 and gRNA2 targets in transgenic plants expressing sgRNA1:Cas9, sgRNA2:Cas9, PTG6:Cas9, or PTG7:Cas9 was examined at T0 generation. Among T0 generation of sgRNA1:Cas9 (n = 32) and sgRNA2:Cas9 plants (n = 20), 44% and 60% of them carried indels whereas 13% and 20% of them had biallelic mutations, respectively (Table 1 and SI Appendix, Fig. S8). By contrast, indels at both targets were detected in all PTG6:Cas9 plants (100%, n = 17) including 35% (gRNA1) and 76% (gRNA2) biallelic mutations (Table 1 and SI Appendix, Fig. S9). However, the chromosomal-fragment deletion between gRNA1 and gRNA2 target sites was not detected in PTG6:Cas9 plants with PCR, which may occur at a lower frequency in regenerated calli/plants than in protoplasts. Nevertheless, the results showed that PTG6 not only expressed two gRNAs simultaneously, but also achieved a higher mutagenesis efficiency at individual targets than sgRNA did (Student’s t test, P < 0.01) (Table 1 and SI Appendix, Figs. S8 and S9).
Table 1.
Targeted mutation efficiency in PTG:Cas9 vs. sgRNA:Cas9 plants
| Gene ID | No. of T0 lines | Editing at gRNA1 target, % | Editing at gRNA2 target, % | Editing at gRNA3/5/7 targets, % | |||
| Mut | Bi-Mut | Mut | Bi-Mut | Mut | Bi-Mut | ||
| sgRNA1 | 32 | 44 | 13 | n.a. | n.a. | n.a. | n.a. |
| sgRNA2 | 20 | n.a. | n.a. | 60 | 20 | n.a. | n.a. |
| PTG6 | 17 | 100** | 35** | 100** | 76** | n.a. | n.a. |
| PTG7 | 17 | 47 | 6 | 100* | 24 | n.a. | n.a. |
| PTG9 | 14 | 86** | 78** | 86** | 57** | 86 | 86 |
The frequency of mutation (Mut) and biallelic mutation (Bi-Mut) at the targeting sites in transgenic lines was examined and calculated based on PCR/RE assays (SI Appendix, Figs. S8–S11). The editing frequencies at gRNA1 and gRNA2 targets were compared between sgRNA and PTG plants. **Student’s t test P < 0.01; *Student’s t test P < 0.05. n.a., not applied.
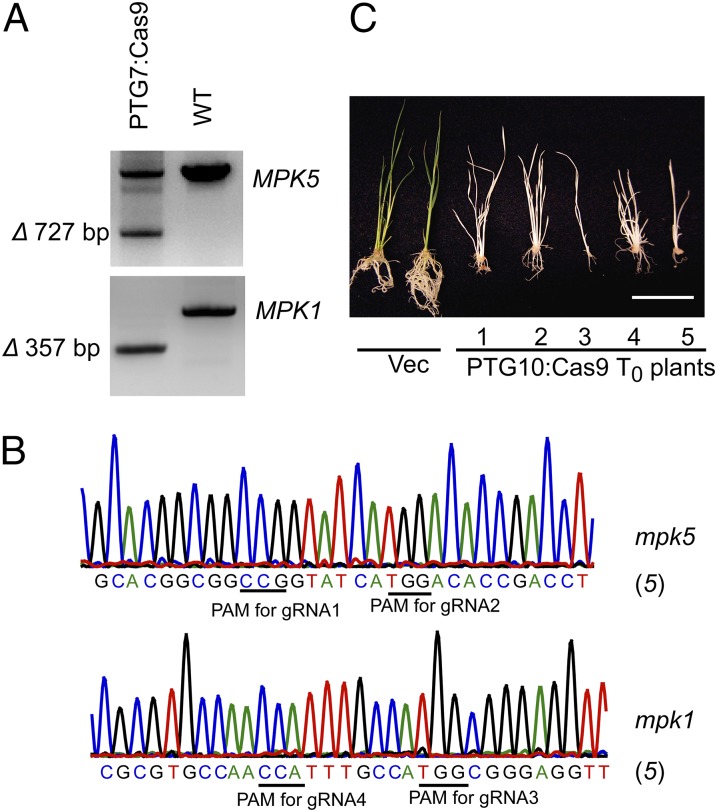
When the total number of targets increased to four in PTG7:Cas9 plants, a comparable indel frequency to sgRNA1:Cas9 plants was observed at the gRNA1 site, but a significantly higher frequency of indels than sgRNA2:Cas9 was found at the gRNA2 site (Table 1 and SI Appendix, Fig. S10). Interestingly, we obtained one PTG7:Cas9 line (6%) carrying biallelic deletion of ∼350 bp in MPK1 and monoallelic deletion of ∼750 bp in MPK5 (Fig. 4 A and B). This result prompted us to further examine the efficiency of all eight gRNAs in PTG9:Cas9 lines. At five gRNA targets (gRNA1/2/3/5/7) whose mutation would destroy RE sites, 50% (n = 14) of PTG9:Cas9 T0 lines carried biallelic mutations at all five targets (SI Appendix, Fig. S11). Interestingly, PCR/RE assays suggested that these five gRNAs exhibited comparable efficiency and that all of them showed significantly higher mutagenesis activities than what we observed in sgRNA1/2:Cas9 lines (Table 1). Sanger sequencing of the fragments from four MPK loci of PTG9:Cas9 plants confirmed that mutations were introduced at all eight sites (SI Appendix, Fig. S12). However, the fragment deletion was detected only at MPK2 and MPK5 loci in two PTG9:Cas9 lines (SI Appendix, Fig. S11). In comparison with protoplasts, the efficiency of targeted chromosomal fragment deletion between paired gRNA/Cas9 cut sites is lower in transgenic plants, which might be due to a relatively lower expression of gRNAs and Cas9 in rice calli/regenerated plants (typically only a single copy of a transgene is integrated into the rice genome after the Agrobacterium-mediated transformation). Our data demonstrate that the PTG method not only increases the gRNA number and targeting sites, but also likely enhances mutagenesis efficiency at individual sites, especially when multiple gRNAs are expressed using PTG.
Fig. 4.
Highly efficient targeted mutagenesis in transgenic rice expressing PTG:Cas9. (A and B) Chromosomal fragment deletion in PTG7:Cas9 plant at T0 generation. Of note, only mpk1 with 358-bp deletion (Δ358) was detected in genomic DNA. Sequence analysis of the PCR products (the number in parentheses) reveals an identical deletion pattern in the transgenic plant. (C) Albino seedlings were regenerated from calli transformed with PTG10:Cas9. Most T0 seedlings (87%, n = 15) exhibited a similar photo-bleach phenotype, suggesting a very high efficiency of knocking out PDS with PTG10:Cas9. Vec, control plants transformed with empty vector. (Scale bar: 5 cm.)
To further substantiate the high efficiency of mutagenesis with PTG/Cas9, we synthesized PTG10 with two gRNAs (gRNA9 and gRNA10) to target the rice phytoene desaturase (PDS) gene (SI Appendix, Fig. S13A). PDS is frequently used as a convenient gene target to examine knock-out efficiency because plants with null functional PDS would lead to a visible photo-bleached phenotype. Strikingly, all PTG10:Cas9 transgenic seedlings (T0 generation, n = 15) regenerated from calli showed photo-bleaching phenotype, and 13 of them were completely albino, indicating that these plants carried null functional PDS (Fig. 4C). Although we identified only one line carrying chromosomal deletions between two gRNA-targeted sites, sequencing of the PDS targets from all of the lines confirmed that indels were introduced at the target sites (SI Appendix, Fig. S13). By comparing these data with the previously reported efficiency of the sgRNA/Cas9 system (9, 11, 29–32), to our knowledge, the PTG/Cas9 approach yielded the highest efficiency of targeted mutagenesis (up to 100%) (Fig. 4 and Table 1) in plants. Our results also demonstrate that targeting one gene with two gRNAs using PTG will greatly increase the efficiency of complete gene knock-out.
Discussion
We developed a general strategy and platform to produce multiple gRNAs from a single synthetic PTG gene by hijacking the endogenous tRNA-processing system (Fig. 1). We also provided a framework to design, synthesize, and use PTG for multiplex genome editing with Cas9. These PTGs were expressed with Pol III promoters (e.g., U3p) in the same manner as sgRNA genes but were not obligated to start with a specific nucleotide (Fig. 2). As a result, current CRISPR/Cas9 vectors for expressing sgRNA could be directly used to express PTG for multiplex genome editing as we demonstrated in this study.
By producing multiple gRNAs from a single polycistronic gene, the PTG technology could be used to improve simultaneous mutagenesis of multiple genomic loci or deletion of short chromosomal fragments (Figs. 3 and 4). Such a genome engineering approach may lead to simultaneous knock-out of multiple protein coding genes or deletion of noncoding RNA regions and other genetic elements. In addition to targeted mutagenesis/deletion, the PTG approach could facilitate other Cas9-based applications in which multiple gRNAs are required. For example, PTG could be used with Cas9 nickase to improve targeting fidelity (13, 33, 34), or with deactivated Cas9 transcriptional-activator or -repressor to manipulate multiple gene expression (35, 36). Given the high processing accuracy and capability of RNase P and RNase Z that we observed (Fig. 2), the tRNA-processing system also could be used as a general platform to produce other regulatory RNAs (e.g., short hairpin RNA or artificial microRNA) from a single synthetic gene. These different classes of regulatory RNAs, like gRNA and short hairpin RNA, also could be compacted into a single polycistronic gene to develop more sophisticated devices for genetic engineering.
Recently, polycistronic transcripts that fused gRNA with a 28-nt RNA (referred to as gRNA-28nt) were successfully used to guide Cas9 to target up to four targets in human cells (12, 13). The synthetic gene with a gRNA-28nt architecture produced mature gRNAs with a 28-nt extra 3′ sequence and also required coexpressing the endonuclease Csy4 from Pseudomonas aeruginosa to cleave the transcript. In comparison with the gRNA-28nt strategy, our approach uses a robust endogenous tRNA-processing system that enables precise production of gRNAs with only a 1- to 4-nt extra sequence at the gRNA 3′ end (Figs. 1 and 2) and carries no additional risk of endonuclease Csy4 toxicity to recipients. Given the extremely large number of tRNA genes with variable sequences and the fact that RNase P and RNase Z precisely recognize RNA substrates with tRNA-like structures (18, 37), there are many choices of tRNA sequences to be embedded in PTG. Furthermore, the tRNA-processing system is universal in all living organisms; thus, the PTG technology could be directly adapted to other organisms for Cas9-mediated genome engineering.
When multiple double-strand breaks (DSBs) in genomic DNA were generated by PTG/Cas9 in rice plants, indels resulting from error-prone NHEJ repairing occurred more frequently than fragment deletions generated by directly joining two DSBs (SI Appendix, Figs. S10 and S11). To date, the molecular mechanism by which two DSBs directly link together to generate chromosomal translocation or fragment deletion in vivo is largely unclear. We speculate that the process leading to such a chromosomal disorder may require two DSBs at the same time interval and is likely determined by the highly dynamic interaction between gRNA/Cas9 cutting and endogenous DNA repairing and also by the distance between DSBs. Due to the differences in the delivery, expression, and activity of gRNAs and Cas9, it is not surprising to see some discrepancies in fragment-deletion frequency between protoplasts (Fig. 3B) and stable transgenic plants and among different PTG transgenic lines (Fig. 4A and SI Appendix, Figs. S9–S11). Because the PTG technology enables the generation of many DSBs in genomic DNAs, it may provide an efficient tool to help dissect the molecular process of chromosomal deletion. More importantly, the PTG technology significantly improves multiplex editing capability and efficiency and is expected to facilitate more sophisticated Cas9 applications, such as targeted mutagenesis and deletion of redundant genes or genetic elements, transcriptional modulation of multiple genes and pathways, modification and labeling of numerous genomic sites, site-specific integration, and gene replacement.
Materials and Methods
Plant Materials.
Rice (Oryza sativa L. ssp) cultivars Kitaake and Nipponbare were used in this study. Rice plants were grown in a green house or growth chamber at 28 °C day/23 °C night with 12 h of light.
Plasmid Vector Construction.
The plasmid vector pRGE32 was used to transiently express U3p:sgRNA or U3p:PTG along with UBIp:Cas9 in rice protoplasts, and pRGEB32 was used for the Agrobacterium-mediated rice transformation (SI Appendix, Fig. S2). See SI Appendix, SI Methods for details about plasmid vector construction.
sgRNA:Cas9 and PTG:Cas9 Expression Constructs.
The U3p:sgRNA1 and U3p:sgRNA2 constructs were made as described previously (24). The specific spacer sequences for gRNA3-gRNA8 (SI Appendix, Table S3) were selected using the CRISPR-PLANT database (www.genome.arizona.edu/crispr/) (38). The PTG genes and U3p:PTG constructs were generated as described in SI Appendix, SI Methods, Figs. S6 and S7, and Table S3. The synthesized PTGs were inserted into the BsaI-digested pRGE32 or pRGEB32 for transient protoplast expression or stable rice transformation, respectively. The sequences of sgRNA1, sgRNA2, and PTG genes used in this study are listed in SI Appendix, Table S1.
Rice Protoplast Transfection.
The rice protoplast preparation and transfection were performed as described previously (24). Briefly, 20 µg of plasmid DNA was used to transfect 2 × 105 protoplasts with a transfection efficiency of ∼40–50%. Total rice genomic DNAs were extracted from the protoplast samples at 36 h after transfection and then used for PCR and sequence analysis.
Agrobacterium-Mediated Rice Transformation.
Transgenic rice plants were generated by the Agrobacterium tumefaciens-mediated transformation using rice mature seed-derived calli according to a conventional protocol (39).
Genomic DNA Extraction and PCR/RE Assay.
Rice genomic DNA was extracted as described previously with the hexadecyltrimethylammonium bromide method (24). To detect mutagenesis at desired sites, target regions were amplified with specific primers (see SI Appendix, Table S2 for primer sequences) using GoTaq DNA polymerase (Promega). The PCR product was separated in 1% agarose gel and stained with ethidium bromide to detect chromosomal-fragment deletion. To detect indels at specific sites with PCR/RE, the PCR products encompassing the target sites were digested with appropriate restriction enzymes (RE) for 5 h and then were analyzed with 1% agarose gel electrophoresis and ethidium bromide staining. The stained gels were imaged using the Gel Doc XRS system (Bio-Rad) and quantified with the Quantity One 4.6 program (Bio-Rad). Selected PCR products were cloned into pGEM-T Easy Vector (Promega) for DNA sequencing.
RNA Extraction, cRT-PCR, and Quantitative PCR.
See SI Appendix, SI Methods for details.
Gene Accession Number in GenBank.
Genes and their GenBank RefSeq accession numbers are as follow: MPK1, Os06g0154500; MPK2, Os08g0157000; MPK5, Os03g0285800; MPK6, Os10g0533600; UBI, Os02g0161900; and PDS, Os03g0184000.
Supplementary Material
Acknowledgments
This work was supported by the Pennsylvania State University and the National Science Foundation Plant Genome Research Program.
Footnotes
Conflict of interest statement: The Pennsylvania State University filed a provisional patent application related to this study.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420294112/-/DCSupplemental.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JF, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 12.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54(4):698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24(17):1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiffer S, Rösch S, Marchfelder A. Assigning a function to a conserved group of proteins: The tRNA 3′-processing enzymes. EMBO J. 2002;21(11):2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutmann B, Gobert A, Giegé P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012;26(10):1022–1027. doi: 10.1101/gad.189514.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbezier N, et al. Processing of a dicistronic tRNA-snoRNA precursor: combined analysis in vitro and in vivo reveals alternate pathways and coupling to assembly of snoRNP. Plant Physiol. 2009;150(3):1598–1610. doi: 10.1104/pp.109.137968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canino G, et al. Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 2009;150(3):1494–1502. doi: 10.1104/pp.109.137950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruszka K, et al. Plant dicistronic tRNA-snoRNA genes: A new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 2003;22(3):621–632. doi: 10.1093/emboj/cdg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakajima N, Ozeki H, Shimura Y. Organization and structure of an E. coli tRNA operon containing seven tRNA genes. Cell. 1981;23(1):239–249. doi: 10.1016/0092-8674(81)90288-9. [DOI] [PubMed] [Google Scholar]
- 21.Nakaar V, Dare AO, Hong D, Ullu E, Tschudi C. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol Cell Biol. 1994;14(10):6736–6742. doi: 10.1128/mcb.14.10.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White RJ. Transcription by RNA polymerase III: More complex than we thought. Nat Rev Genet. 2011;12(7):459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 23.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6(6):1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 25.Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn J, Binder S. RT-PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res. 2002;30(2):439–446. doi: 10.1093/nar/30.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokobori S, Pääbo S. Transfer RNA editing in land snail mitochondria. Proc Natl Acad Sci USA. 1995;92(22):10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE. 2008;3(11):e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(8):691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 30.Feng Z, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(12):4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12(6):797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 33.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249(4970):783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 38.Xie K, Zhang J, Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7(5):923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- 39.Hiei Y, Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc. 2008;3(5):824–834. doi: 10.1038/nprot.2008.46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.