Abstract
The Gene Expression Database (GXD) is an extensive and freely available community resource of mouse developmental expression data. GXD curates and integrates expression data from the literature, via electronic data submissions, and by collaborations with large-scale projects. As an integral component of the Mouse Genome Informatics (MGI) Resource, GXD combines expression data with genetic, functional, phenotypic and disease-related data, and provides tools for the research community to search for and analyze expression data in this larger context. Recent enhancements include: an interactive browser to navigate the mouse developmental anatomy and find expression data for specific anatomical structures; the capability to search for expression data of genes located in specific genomic regions, supporting the identification of disease candidate genes; a summary displaying all the expression images that meet specified search criteria; interactive matrix views that provide overviews of spatio-temporal expression patterns (Tissue × Stage Matrix) and enable the comparison of expression patterns between genes (Tissue × Gene Matrix); data zoom and filter utilities to iteratively refine summary displays and data sets; and gene-based links to expression data from other model organisms, such as chicken, Xenopus and zebrafish, fostering comparative expression analysis for species that are highly relevant for developmental research.
Keywords: embryo, development, resource, bioinformatics, data integration, literature curation
Introduction to GXD
As a mammalian model organism, the mouse is heavily used for developmental research. Tissues from all developmental stages and from many different mouse strains and mutants are subject to detailed expression studies in order to gain insights into the function of genes and the molecular mechanisms that underlie human development, differentiation, and disease. However, these expression data are voluminous, complex, and heterogeneous. They are generated by many different laboratories and scattered through thousands of publications. Without the help of centralized databases, it is impossible to keep abreast of all this information, let alone to access and search these data in an integrated way.
For many years, the Gene Expression Database for Mouse Development (GXD) has provided this critical and unique function to the research community. GXD systematically collects and integrates mouse developmental expression data through curation of the literature, via electronic submission, and by collaboration with large-scale projects. Focusing on endogenous gene expression during development in wild-type and mutant mice and covering data from RNA in situ hybridization, in situ reporter (knock-in), immunohistochemistry, RT-PCR, Northern blot and Western blot experiments, the database has grown tremendously in recent years in terms of data content and search and display utilities (Finger et al. 2011; Smith et al. 2014a). Furthermore, as an integral component of the larger Mouse Genome Informatics (MGI) resource, GXD combines its expression data with other genetic, functional, phenotypic, and disease-oriented data, thereby enabling users to search for expression data and images in many different ways, using a variety of biologically and biomedically relevant parameters (Smith et al., 2014b; Eppig et al., 2015). Here, we briefly describe our data acquisition, curation, and integration efforts, as well as some of GXD’s previously established search utilities. Then we highlight recently added search and display features and describe how best to use them.
Data Curation and Integration
Curators survey journals to identify articles reporting on the types of data GXD collects. The number of publications with embryonic mouse expression information has averaged more than 1,000 per year for the past five years. In a first curation step, GXD curators index expression data in the text of these publications, including supplemental information. The index process identifies the genes whose expression has been studied, the methods used, and the ages analyzed. These annotations are then combined with bibliographic information from PubMed to create the Gene Expression Literature Index. This Index is complete from 1993 with 23,148 references studying 15,309 genes. It is searchable via the Gene Expression Literature Query form (http://www.informatics.jax.org/gxdlit); this search form enables researchers to quickly find publications with specific expression data content.
The index records described above also allow GXD curators to prioritize publications for further annotation. The detailed curation of expression data begins with the entry of probe and antibody information, visualization method, and specimen genetic background and preparation (fixation and embedding material). Curators then record the authors’ descriptions of the results: where expression was observed and, as importantly, where it was absent; how strong or weak the staining; and any pattern of staining, such as whether it was scattered, uniform, or restricted to a part of the tissue.
GXD also integrates and preserves expression data from large-scale projects. GXD has collaborated with the Eurexpress, GenePaint, BGEM, and GUDMAP projects to incorporate their RNA in situ data, as well as some immunohistochemistry data (Diez-Roux et al., 2011; Harding et al., 2011; Magdaleno et al., 2006; Visel et al., 2004). After these projects have ended and their web sites are no longer updated or available, GXD continues to maintain these data and related data connections, such as probe-to-gene associations that might change with new genome builds. The data are fully integrated with all the other data in GXD and MGI and thus readily accessible to many types of searches.
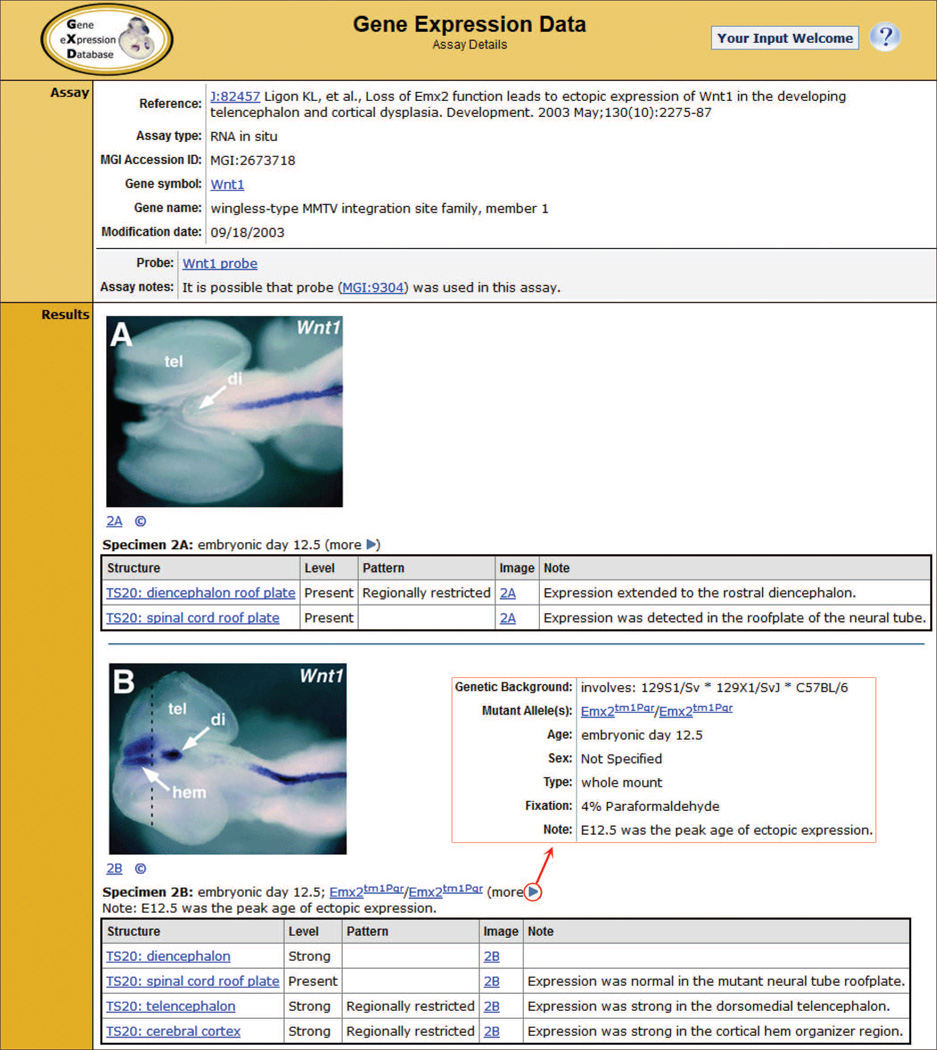
All the expression data in GXD, whether they come from the literature, electronic data submissions, or large-scale projects, are reviewed and recorded in standardized ways following GXD curatorial policy. An example of a detailed assay entry is shown in Figure 1. Annotation of probes, genes, specimens, experimental parameters, and results use standard nomenclature, controlled vocabularies and ontologies. The time and space of gene expression is described using an extensive, hierarchically structured mouse developmental anatomy ontology, developed in collaboration with the Edinburgh Mouse Atlas Project (EMAP) (Hayamizu et al., 2013). Therefore, expression results from different methods with differing spatial resolution can be annotated in a consistent format at the appropriate levels of tissue granularity. Result annotations are displayed together with images of original expression data (along with publisher’s copyright permissions) so that researchers can see and interpret the primary data themselves.
Figure 1.
GXD Assay Detail Page. Annotations from publications (or electronic submissions) are grouped into assays defined by a particular gene, assay type, and probe combination. The assay shown is a comparison of expression patterns between wild type (Specimen 2A) and Emx2 mutant (Specimen 2B) embryos. Results for a particular specimen are annotated using a hierarchically-organized anatomy ontology. Additional controlled vocabularies are used to indicate the expression level and pattern as described by the author. Text notes add further details. The ‘more’ toggle (circled in red) expands to display specimen details, such as the genetic background of the mutant allele, the fixation method and the specimen type (red box). Annotations are accompanied by corresponding images. These images link to the image detail page showing the full figure (with publisher copyright permission).
Through its dedicated curation efforts, GXD has acquired a large amount of expression data. Currently, GXD contains more than 69,000 assay records covering expression data for almost 14,000 genes. These assays include over 268,000 images and nearly 1.5 million annotated expression results. In addition to data from different strains of wild-type mice, GXD has expression data for 2,142 mouse mutants.
The standardized annotations of expression data are key to data integration within GXD and the larger MGI resource. For instance, integration at the gene level associates expression data with Gene Ontology and OMIM disease terms and other gene-based information (Amberger et al., 2015; Gene Ontology Consortium, 2013). Integration at the probe level relates assays targeting the same isoforms and provides links to sequence information. Integration at the tissue level aggregates all the expression data for a specific anatomical structure. Integration at the allele level combines the expression data and phenotypic data for a specific mutant. All of these points of integration enable unique and powerful search capabilities, alleviating the need to search multiple resources for specific pieces of information.
Queries in GXD
The GXD Home page at http://www.informatics.jax.org/expression.shtml is a central starting point for exploring data in GXD. It provides access to all GXD query forms, tutorials on how to find specific types of data, information about current data content, new releases, curatorial policies, collaborators, as well as guidelines for submitting expression data to GXD.
The Mouse Developmental Anatomy Browser (http://www.informatics.jax.org/vocab/gxd/anatomy/EMAPA:16039) lets users explore the developmental anatomy and look up expression data associated with specific anatomical structures and their substructures (Fig. 2). The anatomy browser consists of three sections. The Anatomy Search section provides access to the “abstract” version of the anatomy ontology, referred to as EMAPA, which lists the anatomical structures for all developmental stages (Theiler stages) together with the stage range during which each structure is present (Fig. 2a). Upon selecting a specific term, the Tree View section of the browser displays the corresponding anatomical structure in its hierarchical context (Fig. 2b). Users can navigate through the anatomy by expanding and collapsing branches of the hierarchy, as well as follow links to expression data for specific anatomical structures. The Term Detail section provides additional information about a selected anatomical structure (Fig. 2c). For example, it lists its hierarchical parents. Because the anatomy ontology is organized as a directed acyclic graph (DAG), one anatomical structure can have more than one parent. For example, “amnion ectoderm” is part of the “amnion”, as well as a subclass of “extraembryonic ectoderm”. This enables viewing and searching the anatomy from different perspectives. Upon clicking on another parent term, the corresponding hierarchical organization is shown in the Tree View. The Term Detail section also lets users toggle between the abstract version (showing the range of stages over which the structure exists) and stage-specific versions of the hierarchy (derived from the information in the abstract ontology and referred to as EMAPS) that are then displayed in the Tree View section (Fig. 2d). Stage-specific anatomical terms (EMAPS terms) displayed in the tree view lead to the expression data for the anatomical structure at the specific developmental stage, whereas ‘abstract’ anatomical terms (EMAPA terms) lead to expression data for the anatomical structure at all developmental stages during which it is present.
Figure 2.
Mouse Developmental Anatomy Browser. (a) The Anatomy Search (left) finds anatomy terms containing the search term. The search returns matches in the ‘abstract’ version of the ontology (EMAPA), as indicated by the Theiler stage (TS) ranges beside each term. In the example, amnion ectoderm was selected (red circle). (b) Selecting a term from the search updates the Anatomical Tree View (bottom right). When the tree view is labeled ‘All Theiler Stages’ (blue arrow) this is the abstract tree. The term is highlighted in gold, and a link to the expression results is alongside. (c) The Anatomical Term Detail (upper right) includes the stage range, term ID, synonyms and parent terms. The ontology is in a directed acyclic graph (DAG) format, which allows a structure to have more than one parent. In the example, amnion ectoderm is a type of extraembryonic ectoderm but is also part of the amnion (red box). (d) The picklist of Theiler stages in the Anatomical Term Detail section enables switching the term from the ‘abstract’ to a stage-specific term.
The Gene Expression Data Query Form (http://www.informatics.jax.org/gxd) best illustrates the querying capabilities of GXD. It allows searches for expression data from the anatomical perspective, but there are other search parameters that can be used in combination. We will describe it in detail, together with resulting interactive data summaries and their links to detailed data entries.
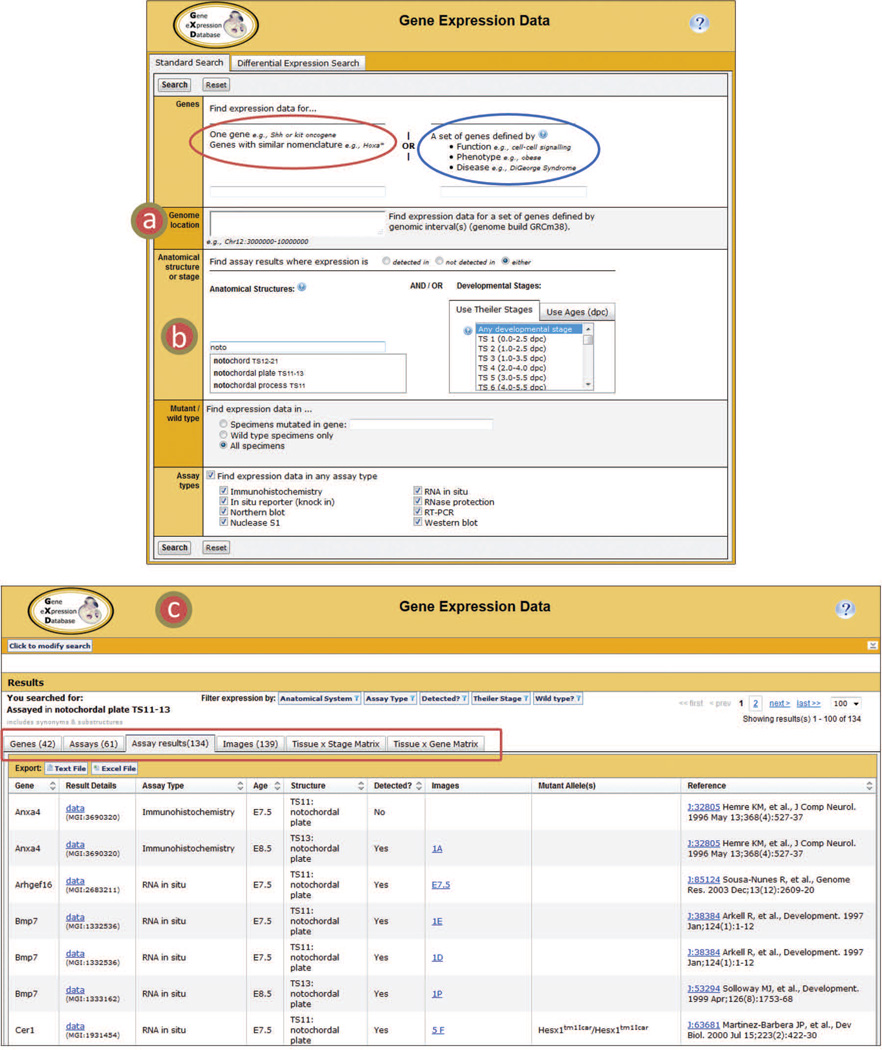
The Standard Search utility (tab) of the Gene Expression Data Query Form allows researchers to formulate questions ranging from simple to complex by either using one search field or by combining several of the available search parameters (Fig. 3). Users can specify a gene or set of genes in expression searches. Gene sets can be specified based on nomenclature, biological function (Gene Ontology (GO) terms; Gene Ontology Consortium, 2013), associated mouse phenotype (Mammalian Phenotype Ontology terms; Smith and Eppig, 2012), or associated human disease (OMIM terms; Amberger et al., 2015) terms. A recently added feature lets researchers define gene sets based on genome location (Fig. 3a) and perform queries such as “Give me all the genes located in a specified chromosomal region and expressed in a specific tissue”, a type of search facilitating the discovery of disease candidate genes.
Figure 3.
Gene Expression Data Query Form and recent enhancements. Users can search for genes based on gene nomenclature (red circle) or on function (Gene Ontology), phenotype (Mammalian Phenotype Ontology), and disease (OMIM) associations (blue circle). (a) Gene sets can now be defined using the Genome location field. Users can add one or more genomic intervals or list chromosomes to restrict the search to a set of particular genes depending on chromosomal location. (b) As the user types in the Anatomical Structure field the autofill displays matches in the Developmental Anatomy Ontology from which to choose. Additional fields on the query form include Developmental Stage (user can choose Theiler stage or age menus), choice of wild type or mutant specimens (default = all; choosing mutated specimens requires entry of the mutated gene symbol), detected or not detected (default = both), and specification of assay type (default = any). (c) The GXD Summary page returns lists of results matching the user’s search. Summaries include the list of Genes, Assays, Assay Results (default tab), Images, and two Matrix views (boxed in red).
The above-described parameters allow users to select the attributes of genes of interest. Other fields on the query form allow users to search by details of the expression patterns of interest. These include: anatomical structure, developmental stage (by age of Theiler Stage), mutant or wild-type, and assay type. One can select for results where expression was detected or not detected. The Anatomical Structure field enables general searches, like ‘mesenchyme’, while also allowing for specific searches, such as ‘1st branchial arch mesenchyme’. The anatomy search is supported by an auto-fill utility (Fig. 3b). Each anatomical structure in the auto-complete list displays the range of Theiler stages at which the structure exists, greatly improving the user’s ability to discern at what developmental stages they can search or when to restrict their search.
The Differential Expression Search (second tab, not shown) lets users query for genes that are expressed in some anatomical structures but not others and/or in some developmental stages but not others.
Expression searches yield data summaries in multiple formats (Fig. 3c). Users can then select the data in the format that best suits their needs. Users can easily move between tabs for Genes, Assays, Assay Results, Images, and two Matrix Views. Following are descriptions of the newest tabs (the Images summary and Matrix Views) and features available on the GXD summary pages.
The Images Summary
Because interpreting expression data is heavily dependent on the image, it can be preferable to start by scanning through relevant images before reviewing the text annotations and experiment details. Researchers can search for images by using the GXD query form and selecting the Images tab on the returned data summary. The Images tab displays images matching the search parameters (Fig. 4a) in a paginated summary that users can scroll through. The image itself is a link to the image detail page (Fig. 4b), where one can see the image pane in the context of the original figure with links to result details. The Images summary also indicates the assay type and provides a direct link to the result details on the assay detail page (Fig. 4c). If a pane has data from a double- or triple-labeling experiment, all genes and their respective assay types are indicated. The default sort order of the image summary is based on assay type with the images of in situ data preceding images of blots.
Figure 4.
Images Summary. (a) A search from the GXD query form returns all images matching the query. Each pane displayed on the summary is a link to the Image Detail page (b), which displays the full publication figure showing all panes. Each entry on the Images summary lists the gene (or genes, if double-labeled) and the assay type. The Result Details link (red circle) leads to the Assay Detail page (c), which displays the structures examined and the strength and pattern of expression observed, as well as specimen and probe information.
GXD Matrix Views
Prior to the implementation of matrix views, results in GXD could only be reviewed in a list, such as the one provided by the assay results tab. Now users can use the matrix views to explore sets of results interactively, starting with high-level overviews and zooming in to more details.
There are two matrix views: Tissue × Stage Matrix (Fig. 5) and Tissue × Gene Matrix (Fig. 6). The Tissue × Stage Matrix is an overview of the spatial and temporal expression results. Each column corresponds to a Theiler stage, with the gestational age range of each stage included in the header for ease of reference. The Tissue × Gene Matrix allows for a comparison of expression patterns among a set of genes. Here the columns correspond to genes.
Figure 5.
Tissue × Stage Matrix. The view displays a set of expression data (in this case expression data for genes associated with the phenotype ‘embryonic lethality’) in a grid comprised of rows of anatomical structures and columns of Theiler stages. (a) The anatomy has toggles (small blue triangles) that can be opened and closed to expand the anatomy to more granular structures (red arrow follows the path from ‘germ layer’ to ‘yolk sac endoderm’). (b) The Matrix Legend details the meanings for the colors and other symbols, such as the red corner that indicates conflicting results (both present and absent) in the structure at that stage, and the small gray circle indicating a tissue exists at the stage, but no annotations are available. (c) When no ‘present’ annotations exist for a term, the matrix displays gold triangles to point users to ‘absent’ annotations in substructures (black bracket). (d) Clicking an individual cell opens a dialog box detailing the results associated with that tissue/stage combination. Users have the option to view the results or associated images in more detail.
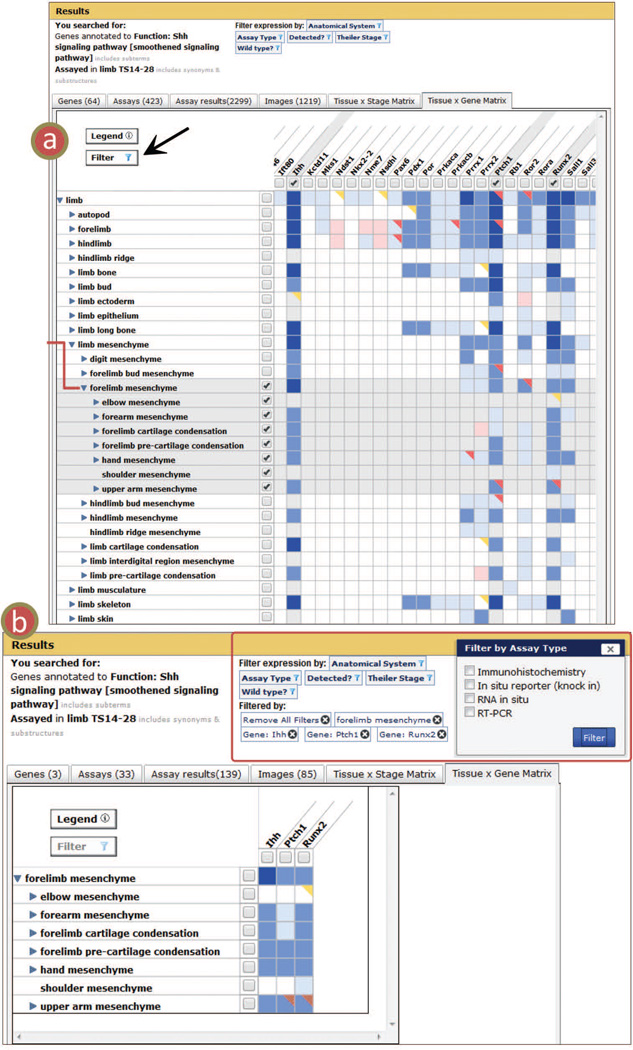
Figure 6.
Tissue × Gene Matrix. View displays a set of expression data in a grid with rows of anatomical structures and columns of genes. (a) The matrix enables comparison of patterns of gene expression such as, in this case, the expression of ‘Shh signaling pathway genes’ in the ‘limb’. To filter the data to generate the image shown in (b) select structures and/or genes with the checkboxes in the row/column headers, then click the filter button (black arrow). (b) Applied filters are listed at the top of the page (red box). Each can be individually removed or all can be removed at once. Filters are applied across all the tabs as evidenced by the drop in Assay Results from 2,299 (tab header in a) to 139 (tab header in b). Additional filters at the top of the page can be used when viewing any of the tab summaries. In (b) the Assay Type Filter is shown open; the four assay types represented in the dataset are listed.
The y-axis of both views displays the anatomy hierarchy. Users can open and close portions of the hierarchy by using the toggles available alongside each structure (Fig. 5a). Opening of each toggle displays successively more granular structures that have been annotated with expression results. In contrast to the Mouse Developmental Anatomy Browser, only those parts of the anatomy relevant to the summarized expression results are displayed. This makes the matrix easier to review.
Matrix Colors
A variety of colors and symbols are used to convey information in the matrix, as detailed in the Matrix Legend (Fig. 5b). Annotations to present are indicated with shades of blue, while annotations to absent are indicated with shades of red. The intensity of the staining indicates the number of annotations to a particular structure, giving an indication of the level of support for the assertion. It should be noted that the logic of managing ‘present’ and ‘absent’ results differs in the matrix anatomy hierarchy, reflecting differences in biological meaning. A ‘present’ annotation to a substructure means that one can infer that gene is expressed in its parent structure as well. For instance, a ‘present’ annotation for ‘embryo endoderm’ also means that the gene is expressed in the parent structure, ‘endoderm,’ as well as the grandparent structure, ‘germ layer.’ The same is not true for an ‘absent’ result. For example, an ‘absent’ annotation for ‘yolk sac endoderm’ does not mean that there is no expression in the parent structure, ‘extraembryonic endoderm.’ Therefore, negative expression results are not projected up the anatomy hierarchy. Matrix cells that display a gold triangle indicate that there are no annotations directly to that structure and that annotations to its substructures are either ‘absent’ (or ‘ambiguous’). This is illustrated in Fig. 5c. Starting with the gold triangle for ‘TS23: endoderm’ and following the trail of gold triangles allows the user to find the ‘absent’ annotations (pink cell) in the more granular structure, ‘yolk sac endoderm.’
Additional Matrix Symbols
There are several other indicators in the Matrix views (Figs. 5,6). Individual cells within the matrix can include both ‘present’ and ‘absent’ results. Conflicting results are indicated with a red triangle in an otherwise blue cell. There might be conflicting annotations because the data set includes results from both wild-type and mutant specimens, includes assay types that have different levels of sensitivity, or because different laboratories produced conflicting results.
Each structure has a range of Theiler stages over which the structure has been shown to exist. However, GXD may not have data for all the stages. In the Tissue × Stage Matrix a light gray circle indicates that the structure exists but GXD has no annotations for it. A blank cell in this matrix means the tissue does not exist at that stage. Indicating a structure’s range helps users see whether expression has been examined early or late in its development. On the Tissue × Gene Matrix, blank cells mean there are no annotations in GXD for that gene in that tissue, either because those experiments have not been published or because they have not been entered into GXD.
Accessing the Matrix Views
Both the Tissue × Stage and Tissue × Gene matrices are available with searches from the GXD Query form. A user can compose a search using any of the search parameters on the GXD query form, e.g. by defining a set of genes using Disease (OMIM) or Mammalian Phenotype Ontology terms. If the search does not include an anatomical structure, the hierarchical anatomy view of the Matrix will begin at the highest level of the anatomy (Fig. 5). Users can then explore the anatomy to the granularity of the results or their interest. Alternatively, if the search specifies an anatomical structure, Matrix views begin at that structure (Fig. 6a). From that point, the user can explore further substructures, but cannot move up the anatomical tree to more general structures. Finally, the GXD homepage has a link to the Gene Expression Data Matrix, a Tissue × Stage Matrix showing all of the data in GXD. This gives users the opportunity to explore all the data interactively in this new matrix view, as well as a mechanism for selecting multiple anatomical systems easily.
After the Matrix…
Once users have identified results in the Matrix that they may wish to view in more detail, clicking on an individual cell provides more information (Fig. 5d). The dialog window that opens identifies the cell by Tissue and either Stage or Gene, indicates the number of genes (on the Tissue × Stage Matrix), and the number of present, absent or ambiguous results. The user has the option to view those results in the Assay Results tab, or the corresponding images in the Images tab. From any of the summaries, the user can proceed to the detailed expression entries.
Filtering
GXD expression summaries feature a variety of filters to manage the search results. These filters are located above the GXD summary tabs and include Anatomical System, Assay Type, Detected?, Theiler Stage, and Wild type (Fig. 6b). Clicking any of these buttons opens a list featuring options that correspond to the returned dataset. The user can, for instance, choose to narrow the results to a specific assay type or remove mutant data. The Anatomical System filter lists high-level vocabulary anatomy terms, and the Detected? option allows the user to choose results that are present (yes) or absent (no). Filtering will propagate across all of the tabs. Indicators of each filter are included in the header, and each filter choice has its own option for removal, so they can be added and removed easily.
Additional filters are found on the Matrix views. Each row and column has checkboxes (Fig. 6a). Column checkboxes allow the user to restrict the Theiler stage (Tissue × Stage Matrix) or the gene (Tissue × Gene Matrix) in the result set. Checkboxes for each row allow the user to restrict the anatomical structures. When a user selects a parent structure, substructures are automatically selected as well because their annotated results are returned in the search. When checked, the row/column becomes highlighted gray to indicate it has been selected. Once the Filter button has been clicked the Matrix becomes restricted to the selections (Fig. 6b). Matrix filters, like other filters discussed above, restrict results across all summary tabs. They are also listed above the tabs and can be removed in the same fashion as the other filters.
Exporting data
The summaries provided by the Genes and Assay Results tab can be downloaded in tab-delimited and Excel™ based formats and exported to other applications. In addition, the genes list (Genes tab) can be forwarded to the MGI Batch Query tool and to MouseMine (see below) for further analysis.
Accessing expression information via the Gene Detail page
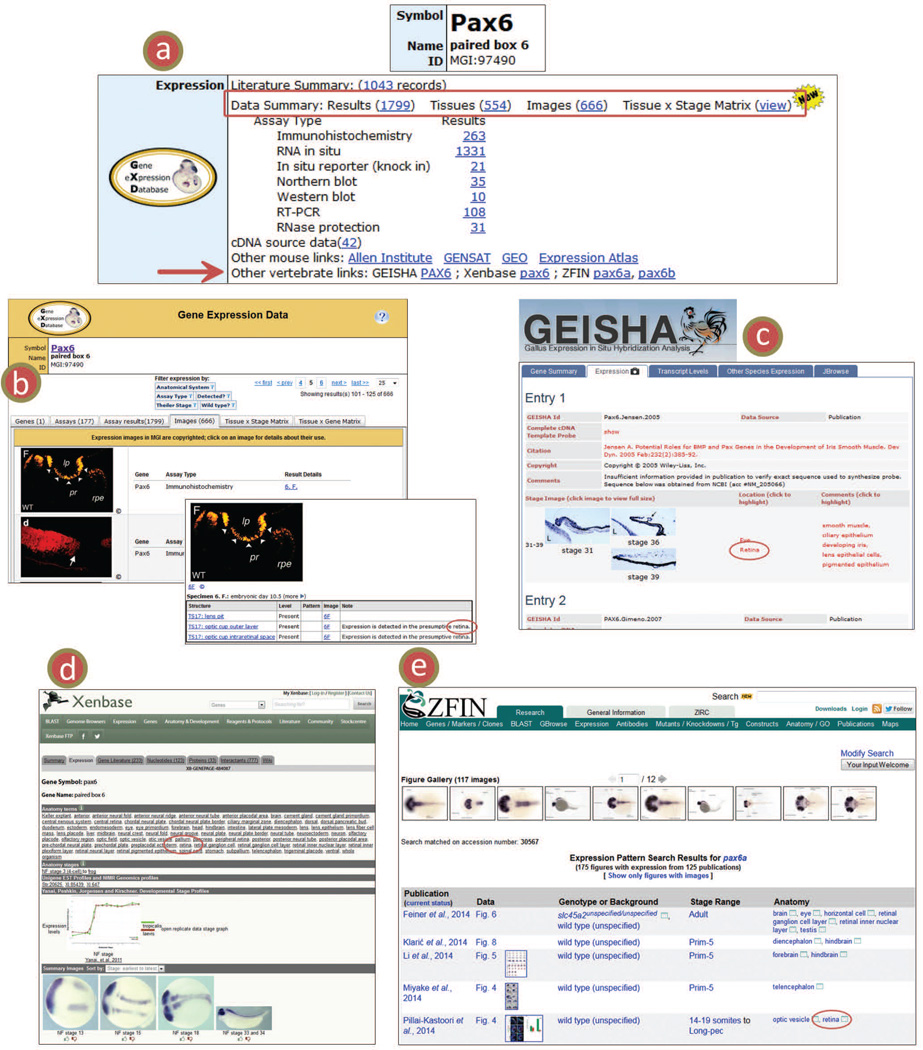
For users who just want to look up expression information for one particular gene, it might be most effective to use MGI’s quick search, accessible in the upper right corner of all MGI pages. Search for a gene symbol or name, and then choose the gene symbol in the resulting list to access its MGI gene detail page. Divided into topic sections, the gene detail page lists all information for that gene in MGI. Gene expression information is summarized in the Expression section of the page (Fig. 7a). There are direct links to Results and Image summaries, as well as a link to the Tissue × Stage Matrix for that gene. There is also a link to the literature summary records of publications that examine expression of that gene.
Figure 7.
Comparing expression across species. (a) The Expression ribbon of the MGI Gene Detail Page (Pax6 shown). Links to data summaries are available for Results, Images, and the Tissue × Stage Matrix (red box). The Other vertebrate links (red arrow) provide access to expression summaries at other model organism databases. The links list the database name then the gene symbol. In those cases where a gene duplication event has occurred, GXD links to both paralogs (note pax6a, pax6b for ZFIN). (c–e) The link opens a separate browser tab showing the expression summary for that gene at the model organism database. Compare expression in mouse, viewed in the GXD Images Summary for Pax6 (b), to expression in chicken at the GEISHA resource (c), Xenopus at the Xenbase resource (d), and zebrafish at the ZFIN resource (e). Note that all four resources have recorded Pax6 expression in retina (red circles).
Comparing gene expression across different model organisms
Comparison of expression patterns among different organisms can provide crucial insights in developmental processes. To make these comparisons easier, GXD has recently implemented gene-based links to expression data within GEISHA (Gallus Expression in Situ Hybridization Analysis; Darnell et al., 2007), ZFIN (Zebrafish Information Network; Bradford et al., 2011), and Xenbase (Xenopus Database; Bowes et al., 2010), which are database resources for other vertebrate model organisms important for developmental research. These links, accessible from the Expression section of the MGI Gene Detail pages (Fig. 7a), are based on gene orthology assertions provided by GEISHA, ZFIN, and Xenbase. They are displayed only if there are expression data for the gene available at these resources.
Batch queries, data reports, and programmatic access
Genomic approaches such as microarray or high-throughput-sequencing experiments often yield lists of “interesting genes” that then need to be examined further. GXD and the larger MGI resource provide several means to effectively obtain information about many genes in a single search. Using the MGI Batch Query tool (http://www.informatics.jax.org/batch), one can enter or upload a list of gene symbols, as well as various ID types (e.g. sequence ID’s, NCBI gene ID’s, etc.), and query for specific information about these genes in MGI. Users can choose to retrieve GXD expression results for each of these genes in web-based, tab delimited, or Excel formats, allowing these data to be exported to other applications for further analysis.
For more powerful querying and reporting needs and to support comprehensive programmatic access, GXD expression data are also available through MGI’s MouseMine (http://www.mousemine.org). Several predefined reports provide for bulk access to expression data via lists of genes, publications, or anatomical structures. A user can customize any report (or create a new one from scratch); download any result in a number of formats (e.g., JSON or tab-delimited); save lists (e.g., lists of genes or publications) and use those lists to drive further querying. With a few clicks, a user/developer can obtain a RESTful URL or working code snippet (in a variety of languages) to run any query programmatically. Underlying all these features is a comprehensive web services API. MouseMine is based on the InterMine framework. InterMine has been described previously (Smith et al. 2012) as well as elsewhere in this volume.
Additional mechanisms of access to data in GXD include our FTP site (ftp://ftp.informatics.jax.org/pub/), custom SQL reports provided upon request via MGI User Support, and online SQL accounts that give programmatic access to a read-only copy of the database (see the MGI developer’s tools page for details on how to access these services, http://www.informatics.jax.org/software.shtml). A database schema browser is available at http://www.informatics.jax.org/schema_pg/. Finally, the entire MGI database, including GXD, is available for download in both PostgreSQL and MySQL formats.
Future Directions
GXD will continue to collect and integrate different types of mouse developmental expression data, including expression data from high-throughput sequencing (HTS) experiments, place these data in the larger genetic, functional, phenotypic, and disease context, and develop search and display tools that let researchers readily access and explore the expression data in this context. Combining expression and interaction data will provide information about where and when molecular interactions occur. Explicit anatomical cross-references between mouse expression and phenotype data will support the correlation of these data, for example by displaying and superimposing both types of data in a common matrix view, and thus facilitate insights into molecular mechanisms of diseases. Anatomical cross-references between model organism databases from different species, currently being established in collaboration with the Uberon project (Haendel et al., 2014), will enable an anatomy-based comparison of expression patterns, in addition to gene-based comparisons, thus fostering our understanding of, and research into, molecular mechanisms of development, differentiation, and disease.
Acknowledgements
We would like to thank the software group, user support, and our colleagues from the other MGI projects for their contributions to the GXD project and to the MGI resource. GXD thanks the GEISHA, Xenbase and ZFIN resources for their help in establishing links between our respective databases. GXD is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health (NIH); Grant number: HD062499.
References
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38(suppl 1):D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Bauer Schaper H, Schaper K, Shao X, Singer A, Sprague J, Sprunger B, Van Slyke C, Westerfield M. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011;39(suppl 1):D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Davey S, Konieczka JH, Yatskievych TA, Antin PB. GEISHA: An in situ hybridization gene expression resource for the chicken embryo. Cytogenet Genome Res. 2007;117:30–35. doi: 10.1159/000103162. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nürnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dollé P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE Mouse Genome Database Group. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2011;39:D835–D841. doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Gene Ontology annotations and resources. Nucleic Acids Res. 2013;41:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendel MA, Balhoff JP, Bastian FB, Blackburn DC, Blake JA, Bradford Y, Comte A, Dahdul WM, Dececchi TA, Druzinsky RE, Hayamizu TF, Ibrahim N, Lewis SE, Mabee PM, Niknejad A, Robinson-Rechavi M, Sereno PC, Mungall CJ. Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon. J Biomed Semantics. 2014;5:21. doi: 10.1186/2041-1480-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y, Sharghi M, Tindal C, McMahon AP, Gottesman B, Little MH, Georgas K, Aronow BJ, Potter SS, Brunskill EW, Southard-Smith EM, Mendelsohn C, Baldock RA, Davies JA, Davidson D. The GUDMAP database – an online resource for genitourinary research. Development. 2011;138:2845–2853. doi: 10.1242/dev.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Wicks MN, Davidson DR, Burger A, Ringwald M, Baldock RA. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J Biomed Semantics. 2013;4:4–15. doi: 10.1186/2041-1480-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, Norland SM, Rice DS, Dosooye N, Shakya S, Mehta P, Curran T. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome. 2012;23:653–668. doi: 10.1007/s00335-012-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RN, Aleksic J, Butano D, Carr A, Contrino S, Hu F, Lyne M, Lyne R, Kalderimis A, Rutherford K, Stepan R, Sullivan J, Wakeling M, Watkins X, Micklem G. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:3163–3165. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Berghout J, Campbell J, Corbani LE, Forthofer KL, Frost PJ, Miers D, Shaw DR, Stone KR, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD: 2014 update. Nucleic Acids Res. 2014a;42:D818–D24. doi: 10.1093/nar/gkt954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Kadin JA, Richardson JE, Ringwald M. The gene expression database for mouse development (GXD): putting developmental expression information at your fingertips. Dev Dyn. 2014b;24:1176–1186. doi: 10.1002/dvdy.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]