Abstract
The lysine-specific histone methyltransferase KMT2D has emerged as one of the most frequently mutated genes in follicular lymphoma (FL) and diffuse large B cell lymphoma (DLBCL). However, the biological consequences of KMT2D mutations on lymphoma development are not known. Here we show that KMT2D functions as a bona fide tumor suppressor and that its genetic ablation in B cells promotes lymphoma development in mice. KMT2D deficiency also delays germinal center (GC) involution, impedes B cell differentiation and class switch recombination (CSR). Integrative genomic analyses indicate that KMT2D affects H3K4 methylation and expression of a specific set of genes including those in the CD40, JAK-STAT, Toll-like receptor, and B cell receptor pathways. Notably, other KMT2D target genes include frequently mutated tumor suppressor genes such as TNFAIP3, SOCS3, and TNFRSF14. Therefore, KMT2D mutations may promote malignant outgrowth by perturbing the expression of tumor suppressor genes that control B cell activating pathways.
Introduction
A majority of B cell lymphomas arise from germinal center (GC) B cells, including follicular lymphoma (FL) and diffuse large B cell lymphoma (DLBCL) - the most common forms of non-Hodgkin lymphoma. Germinal centers (GC) are highly specialized regions within peripheral lymphoid tissues where B lymphocytes undergo rapid proliferation, somatic hypermutation (SHM), Ig class switching, and differentiation, in order to form high affinity antibody secreting cells during an immune response. Extensive GC B cell proliferation coupled to mutagenesis is thought to facilitate the emergence of pro-oncogenic genetic lesions that trigger lymphoma development1. FL arises form GC B cells that have acquired a t(14;18) translocation presumably during earlier VDJ recombination, leading to constitutive expression of the anti-apoptotic BCL2 oncogene2. However, this translocation is also detectable in many healthy adults who never develop the disease2. Additional mutations must therefore contribute to lymphomagenesis. Recent genome sequencing studies have catalogued somatic mutations that may promote GC derived lymphomas3,4. Notably, the lysine methyltransferase KMT2D (also called MLL2 or MLL4) emerged as one of the most frequently mutated genes with reported mutation frequencies ranging from 30% to 85% in both FL and DLBCLs3,4. KMT2D is a catalytic component of the mammalian COMPASS (Complex Of Proteins Associated with Set1) complex, which facilitates transcription through mono- and di-methylation of histone 3 lysine 4 (H3K4) at enhancer regions in DNA5–8. The reported spectrum of KMT2D mutations in lymphoma, with frequent nonsense mutations upstream from the catalytic SET domain and without an apparent mutation hotspot suggests loss of enzymatic function3,4. Here we use mouse models and molecular studies to define KMT2D function in B cells and lymphomagenesis.
Results
KMT2D deficiency promotes lymphoma development in vivo
To directly test the effect of KMT2D deficiency in the development of GC derived lymphoma we utilized the vavP-Bcl2 mouse model. In this model, the vav promoter drives expression of the Bcl2 oncogene in all hematopoietic lineages and this results in the development of B cell lymphomas that recapitulate key aspects of the genetics, pathology, and GC origin of human FLs9–11. To knockdown Kmt2d we transduced unselected vavP-Bcl2 (C57BL/6) transgenic fetal liver cells (ED 14.5) which are a rich source of hematopoietic progenitor cells (HPCs) with the MSCV retrovirus encoding a GFP reporter and short hairpin RNAs targeting Kmt2d (sh-Kmt2d; n=30), empty vector (Vector; n=37) and a retrovirus encoding the Myc oncogene as a positive control (c-Myc; n=16). We injected an unsorted mix of transduced and untransduced HPCs into syngeneic (C57BL/6), wild type, lethally irradiated, female mice and monitored the recipients for 200 days by peripheral blood smear for the emergence of lymphomas (Fig. 1a). Knockdown of Kmt2d caused a significant acceleration of lymphomagenesis and an increase in FL penetrance from 30% to 60% (Fig. 1b). Compared to the unsorted HPCs they derived from and to HPCs transduced with empty retrovirus, the Kmt2d-shRNA-expressing lymphomas displayed a significant enrichment of cells transduced with two different Kmt2d shRNAs tethered to GFP, (Fig. 1c). We confirmed reduction of Kmt2d mRNA levels in mouse B cells expressing the Kmt2d shRNA constructs (Fig. 1d and Supplementary Fig. 1a).
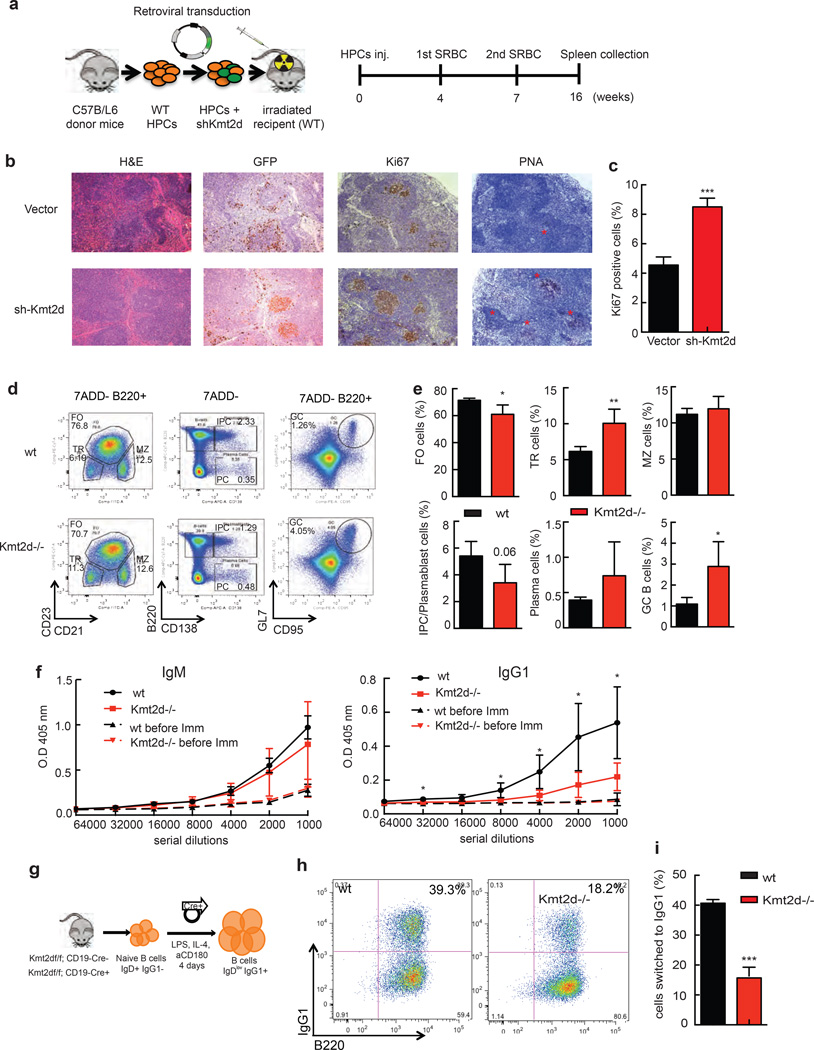
Figure 1. Kmt2d deficiency accelerates B cell lymphoma development in mice.
(a). Diagram of adoptive transfer model of FL based on the vavP-Bcl2 transgenic mouse and retroviral transduction of HPCs followed by reconstitution in lethally irradiated, syngeneic, female mice. (b). Kaplan-Meier curve of C57BL/6 mice transplanted with VavP-Bcl2 HPCs transduced with MSCV-GFP retrovirus (black, n=37, MSCV-GFP encoding shRNAs against Kmt2d (red, n=30), or MSCV-GFP encoding c-Myc (grey, n=16). Statistical significance of survival difference was determined by the log rank test: sh-Kmt2d versus vector: p=0.03; c-Myc versus vector p<0.001). (c). Splenic lymphoma cells of mice that had been injected with VavP-Bcl2 HPCs transduced with retrovirus encoding GFP only or retrovirus co-expressing one of two independent Kmt2d shRNAs (#1 and #2) and GFP were compared by flow cytometry to the same VavP-Bcl2 HPCs prior to injection into mice.. (d). Kmt2d mRNA was quantified qRT-PCR in MACS-sorted B220+ lymphoma B cells from recipients of VavP-Bcl2 HPCs transduced with MSCV-GFP (vector) or MSCV-sh-Kmt2d-GFP (sh-Kmt2d). (e). At pre-terminal stage spleens from indicated recipient mice (vector n=9, shRNA-Kmt2d #1 n=11, c-Myc n=5) were removed and their weight normalized to body weight. Representative images are shown on the right. Values correspond to average ± s.d. Statistical significance in d and e was determined by the two-tailed Student’s t-test: *p<0.05, ***p<0.001. (f). At pre-terminal stage tissue was extracted from recipient mice and stained with H&E, antibody specific for B220, Ki67, PNA or TUNEL. Scale bars indicate 400 µm. (g). At pre-terminal stage whole spleens were isolated and cellular composition analyzed by flow cytometry. Representative images from 4 tumors of each genotype analyzed.
The mice transplanted with vavP-Bcl2-shKmt2d HPCs showed significant splenomegaly and lymphomas were marked by pathognomonic follicular expansion of neoplastic B220+ PNA+ B lymphocytes with low Ki67 staining. Positivity of B cells for PNA and their localization within follicular structures is indicative of their germinal center origin (Fig. 1f). Compared to lymphomas arising in control animals (recipients of vavP-Bcl2 HPCs expressing empty vector) the Kmt2d deficient tumors revealed greater expansion of neoplastic B220+ PNA+ B cells, with advanced destruction of underlying splenic architecture and invasion of the red pulp in nodular and sometime diffuse patterns (Fig. 1f). Kmt2d deficient tumors were composed of a greater number of larger centroblast-like B cells (Supplementary Fig. 1c) and showed more prominent extra-nodal infiltration into the lung, liver, and kidneys (Supplementary Fig. 1d and not shown). Immunophenotyping showed a similar composition of cells in the control and Kmt2d deficient lymphomas with neoplastic B cells expressing B220, CD19, IgM, IgD, and the GC marker GL7 (Fig. 1g and Supplementary Fig. 1b, Supplementary Table 1) PCR analysis of the immunoglobulin light chain (IgL) locus indicated clonal disease (Supplementary Fig. 1e), and sequence analysis of the VDJH4 variable region showed evidence of SHM (Supplementary Fig. 1f). Hence, Kmt2d deficiency cooperates with Bcl2 and promotes the development of high-grade, GC derived FLs.
Next, we analyzed the potential tumor suppressor function of KMT2D in the absence of any cooperating genetic lesions. We crossed Kmt2d conditional KO mice (Kmt2df/f)7 with a CD19-Cre strain to induce Kmt2d deletion in CD19+ early B cells. The majority (58%) of Kmt2df/f CD19-Cre+ mice (herein referred to as Kmt2d−/−) became moribund with a medium survival of 338 days (Supplementary Fig. 1g). Pathology indicated that the Kmt2d−/− B cell lymphomas in spleen and lymph nodes arose from a pre-GC B cell and were composed of monotonous atypical B lymphocytes with high proliferative index (> 90% KI67+) and abundant apoptotic cells by TUNEL (Supplementary Fig. 1h). Flow cytometry revealed CD19+, B220+, IgM+ positive B cells, often expressing Igκ or Igλ+, with variable IgD and CD138 (Supplementary Fig. 1i, Supplementary Table 1). Genomic analyses of the immunoglobulin locus indicated oligoclonal origin from cells that had undergone V(D)J recombination at IgH and Igκ loci (Supplementary Fig. 1k, l). However, the lymphoma cells did not undergo class switch recombination (CSR) and retained germ-line Sµ bands (Supplementary Fig. 1m). They also showed no evidence of somatic hypermutation (SHM) at the IgH locus (Supplementary Fig. 1n), and lacked expression of markers of mouse GC B cells (PNA, (Supplementary Fig. 1h)). Although the mouse tumors may not directly resemble human lymphomas, these results indicate that Kmt2d acts as a tumor suppressor in B lymphocytes and this contrasts with its oncogenic function in the myeloid lineage12.
KMT2D mutations are typically seen in lymphomas that originate form GC B cells which are exposed to the genotoxic activity of the GC specific enzyme activation-induced cytidine deaminase (AID). Therefore we tested if the genomic instability caused by AID would synergize with Kmt2d deficiency to promote lymphoma development in vivo. We crossed the Kmt2df/f CD19-Cre+ mice to animals overexpressing AID (AICDA; AID-Tg) and observed a further acceleration of lymphoma onset (Supplementary Fig. 1g). The Kmt2d−/−; AID-Tg tumors were more aggressive with extensive dissemination into solid organs and complete effacement of the splenic architecture by diffuse proliferation of large atypical B220+ B cells with monotypic expression of IgL light chain and very high proliferative fraction (KI67 positivity >90%). Neoplastic cells were focally positive for the plasmacytic marker CD138 and had accumulation of intracytoplasmic immunoglobulins, suggesting plasmacytic differentiation (Supplementary Fig. 1i, j). These tumors were oligoclonal and contrary to tumors arising in Kmt2d−/− mice, they showed AID-induced CSR and SHM while lacking the GC marker PNA (Supplementary Fig. 1k–n). Hence, AID-induced genomic instability, a hallmark feature of the mutagenic GC environment, cooperates with Kmt2d deficiency in lymphomagenesis.
KMT2D deficiency affects physiological B cell behavior
Heritable nonsense mutations of KMT2D are a major cause of the rare congenital Kabuki syndrome (also known as Kabuki Make-up or Niikawa–Kuroki Syndrome). The syndrome is named for its typical facial features and often comprises a mild immune defect with decreased production of class-switched antibodies and a propensity for ear infections, while a link to tumor development is not clearly established13. We wanted to examine how KMT2D deficiency affects normal B cells. First, we analyzed KMT2D expression by RNAseq across purified mature B cell subsets isolated from human tonsils. KMT2D expression levels were similar in naïve, centroblast, centrocyte, and memory B cells followed by a reduction in plasma B cells, suggesting a functional role for KMT2D before terminal B cell differentiation (Supplementary Fig. 2a). Next, we examined the effect of KMT2D knockdown on GC formation using a transplantation model with wild type HPCs transduced with empty vector or Kmt2d shRNA followed by immunization with sheep red blood cells (Fig. 2a). In control mice, all GCs resolved by week 16 as indicated by loss of PNA and Ki67. In contrast, Kmt2d knockdown mice showed persistent GCs beyond week 16 and consisted of B cells with high PNA and Ki67 staining (Fig. 2b, c). To determine how complete genetic Kmt2d inactivation affects mature B cell populations, we examined Kmt2d−/− mice aged four and five months, without immunization and before lymphoma onset. Flow cytometric analysis of splenocytes harvested from wild type and Kmt2d−/− mice indicated equal numbers of total B220+ B cells, intermediate plasmablasts (IPCs, B220+CD138+), and plasma cells (B220−CD138+) in wild type and Kmt2d−/− mice (Supplementary Fig. 2b, c). We observed a significant two-fold increase in transitional B cells (B220+ CD21− CD23−) and a trend towards elevated germinal center B cells (GC, B220+ GL7+ CD95+) in the Kmt2d−/− mice (Supplementary Fig. 2b–e). Next, we measured the impact of Kmt2d deletion on germinal center formation, six days after immunization. We found a significant but modest decrease in follicular B cells (FO, B220+ CD23+ CD21lo), a trend towards decreased plasmablasts, increased transitional B cells (TR), and most notably a significant three-fold increase in GC B cells in Kmt2d−/− splenocytes compared to wild type (Fig. 2d,e). These results indicate that Kmt2d loss results in expansion in the numbers of GC B-cells after immunization, which represent the cell from which DLBCLs and FLs arise in humans.
Figure 2. KMT2D deficiency affects physiological B cell behavior.
(a). Schematic diagram of SRBC immunization study (SRBC: sheep red blood cell). (b) Representative spleen sections from wild type C57BL/6 females transplanted with HPCs expressing MSCV-GFP or MSCV-shKmt2d shRNA#1 (n=3) and stained with H&E and the indicated markers harvested 16 weeks after SRBC immunization; red stars indicate PNA positive cells. (c). Quantification of Ki67 staining. Values represent mean ± SD (n=3 females per genotype). Two-tailed Student’s t-test was used to determine statistical significance (***p<0.001). (d). Representative FCM analysis of splenocytes harvested from Kmt2d+/+ (wt, 2 males and 2 females; 1.5–2 month old) or Kmt2d−/− mice (3 males and 1 female; 1.5–2 month old) 6 days after NP-CGG immunization. Cells were first gated on live (7ADD−) B220+ lymphocytes to determine percentage of Germinal Center B (GC, CD95+GL7+), transitional B (TR, CD21−CD23−), follicular zone B (FO, CD23+CD21lo), marginal zone B (MZ, CD23lo CD21+), and intermediate plasma cells (IPC, B220+CD138+) cells. Plasma cells (PC, B220−CD138+) cells were gated on live cells (7ADD−). (e). Quantification of flow cytometry data. Values represent mean ± SD (n=4 mice per genotype; wt: 2 males and 2 females, Kmt2d−/−: 3 males and 1 female; 1.5–2 months old). Two-tailed Student’s t-test was used to determine statistical significance (*p<0.05, **p<0.01). Antibodies used are described in methods. (f). NP specific IgM and IgG1 serum levels determined by ELISA from wt or Kmt2d−/− mice before (dashed lines) and 6 days after NP-CGG immunization. The bars show mean ± SD (n=4 mice per genotype; same as in e). Two-tailed Student’s t-test was used to determine statistical significance (*p<0.05). Data correspond to one representative assay from a total of 2 independent assays. (g). Schematic diagram of B cell differentiation assay. (see also Methods) (h). FCM analysis of IgG1 class switch recombination in B220+ cells from wt and Kmt2d−/− mice 96h post-stimulation with LPS, IL-4, and anti-CD180. (i). Quantification of B220+ IgG1+ cells for 2 independent experiments. Values represent mean ± SD (n=5 mice per genotype, 2 females and 3 males, 2.5–5 month old). Two-tailed Student’s t-test was used to determine statistical significance (***p<0.001).
To determine if Kmt2d loss affects B cell antibody production we measured serum IgM and IgG1 levels by ELISA in wild type and Kmt2d−/− mice. Results showed that IgM antibody levels were similar for both groups of mice under basal conditions and while the wild type mice showed the expected increase of IgG1 following NP-CGG immunization, Kmt2d−/− mice had significantly decreased IgG1 levels indicating a class switch defect (Fig. 2f). Consistent with these in vivo findings we also observed a defect in class switch recombination (CSR) to IgG1 in Kmt2d-deficient B cells after in vitro stimulation with lipopolysaccharide (LPS), anti-CD80 and interleukin-4 (IL-4) as indicated by reduced surface IgG1 expression in the Kmt2d−/− B cells (Fig. 2g–i). Hence, KMT2D loss affects B cell differentiation and impedes the B cell immune response in a manner consistent with the mild immune defect associated with Kabuki syndrome.
Consequences of KMT2D mutations in human lymphomas
To explore the effects of KMT2D mutations on clinical behavior, we established the KMT2D mutation status in a cohort of 104 human FL specimens. We detected KMT2D mutations in nearly 40% of samples without an apparent hotspot (Fig. 3a, b; Supplementary Table 2). In these FLs, 38 of 104 samples had KMT2D mutations with 4 being homozygous. Of a total of 49 KMT2D mutations, 36 were nonsense, 12 missense, and 1 frameshift. KMT2D mutations in FL were not significantly associated with FL grade (Supplementary Fig. 3a).
Figure 3. Consequences of KMT2D mutations in human FL and DLBCL.
(a). Percentage of FL (n=104) specimens carrying KMT2D mutations according to type of mutation. (b). Schematic diagram of the KMT2D mutations in FL specimens. (c) and (d). Kaplan-Meier curves representing overall survival (OS)(c) and progression free survival (PFS) (d) in years from DLBCL cases for three groups according to KMT2D mutation status (wt, n=215; nonsense mutation, n=37; missense mutation, n=43). Significance was estimated with the log-rank test. (e). Supervised analysis of the 100 most differentially expressed genes comparing human FL specimens that are KMT2Dwt (n=12) and KMT2Dmut (n=7). Columns represent individual cases, rows correspond to the different genes indicated, the expression values z-scores of rpkm (scaled by row) as shown in the color bar. Columns on the right represent a summary of all mutant and wild type cases. (f). Supervised analysis as in (e) comparing the 100 most differentially expressed mRNAs in MACS-purified mouse B220+ B cells from VavP-Bcl2-vector (Vector, n=4) and VavP-Bcl2-shKmt2d mouse lymphomas (n=5). (g). GSEA of 333 genes significantly down regulated in KMT2Dmut FLs (p-val<0.05) compared to genes ranked log2 fold change in VavP-Bcl2-shKmt2d vs. VavP-Bcl2-vector B220+ lymphoma B cells. (h). GSEA analysis as in (g) of down regulated genes (n=820) in VavP-Bcl2-shKmt2d vs. VavP-Bcl2-vector B220+ B cell lymphomas (p-adj<0.1) compared to genes ranked log2 fold change in KMT2Dwt FLs vs. KMT2Dmut FLs. NES, normalized enrichment score. FDR, false discovery rate. (i). Pathway analysis of down regulated genes (n=347) identified in the GSEA leading edge analyses from (c) and (d) and compared to signatures from the Lymphochip database and MysigDB. The background included around 24,000 genes from Refseq gene annotation. Statistical significance was determined by hypergeometric tests and shown in the color key. The red color indicates (in log10) the over-represented p-values and the blue shows under-representation.
Next, we analyzed KMT2D status in a cohort of 347 newly diagnosed, clinically annotated DLBCL cases, that were all treated with rituximab plus CHOP (R-CHOP: cyclophosphamide, vincristine, doxorubicin, prednisone) at the BC Cancer Agency (Vancouver), and which were classified as GCB or ABC subtypes based on gene expression profiling. The cases were selected on the basis of the following criteria: 16 years of age or older; histologically confirmed de novo DLBCL according to the 2008 WHO classification; available DNA extracted from fresh frozen biopsy material (tumor content >30%). The overall frequency was similar to our FL cohort, however we noticed a significantly higher prevalence of nonsense mutations in the germinal center (GC) subtype (17.6%) compared to the activated B cell (ABC) subtype (8.4%) (Supplementary Fig. 3b). KMT2D mutations were not significantly linked to overall survival (OS), progression-free survival (PFS), disease specific survival (DSS) and time to progression (TTP) (Figs. 3c, d and Supplementary Figs. 3c, d, Supplementary Table 2, see methods for further details). The lack of correlation may indicate no effect on this specific treatment or it may reflect alternate changes in KMT2D wild type tumors that equally affect outcomes.
KMT2D controls a common set of genes in mouse and human FLs
Next we investigated the transcriptional changes related to KMT2D mutation status by RNA-seq on 7 FL specimens with KMT2D nonsense mutations and 12 KMT2D wild type specimens. As expected the most differentially expressed genes in KMT2D nonsense mutant FLs were skewed towards gene down-regulation such that among the top 100 genes 70% were decreased while that fraction decreased to 55% when 500 genes were included (Fig. 3e, Supplementary Figs. 3e, f). Similarly, RNA-seq on MACS purified mouse B220+ cells from Kmt2d knockdown (n=5) and control lymphomas (n=4) revealed that Kmt2d deficient lymphomas had an overall reduction in transcript levels (Fig. 3f, Supplementary Figs. 3g, h). Moreover, genes down regulated in the mouse Kmt2d deficient lymphomas were highly significantly enriched among genes down regulated in human KMT2D mutant specimens and vice versa, Fig. 3g, h, Supplementary Table 3). By contrast there was no enrichment among the up-regulated genes (data not shown). This suggests down regulation of a conserved gene expression signature in human and mouse KMT2D deficient lymphomas. To further explore these signatures we examined the leading edge genes that drive this reciprocal relationship for potential functions perturbed by KMT2D (Supplementary Table 4). The analysis revealed a significant enrichment for genes implicated in the immediate early response, the IL6, IL10, RAS, and TNF signaling pathways (Fig. 2i, Supplementary Table 4), and plasma cell differentiation related genes (Supplementary Fig. 3i, j). Hence, in human and mouse FLs, KMT2D controls a common set of genes related to immune signaling and differentiation pathways.
Epigenetic effects of KMT2D on target genes in mouse and human lymphomas
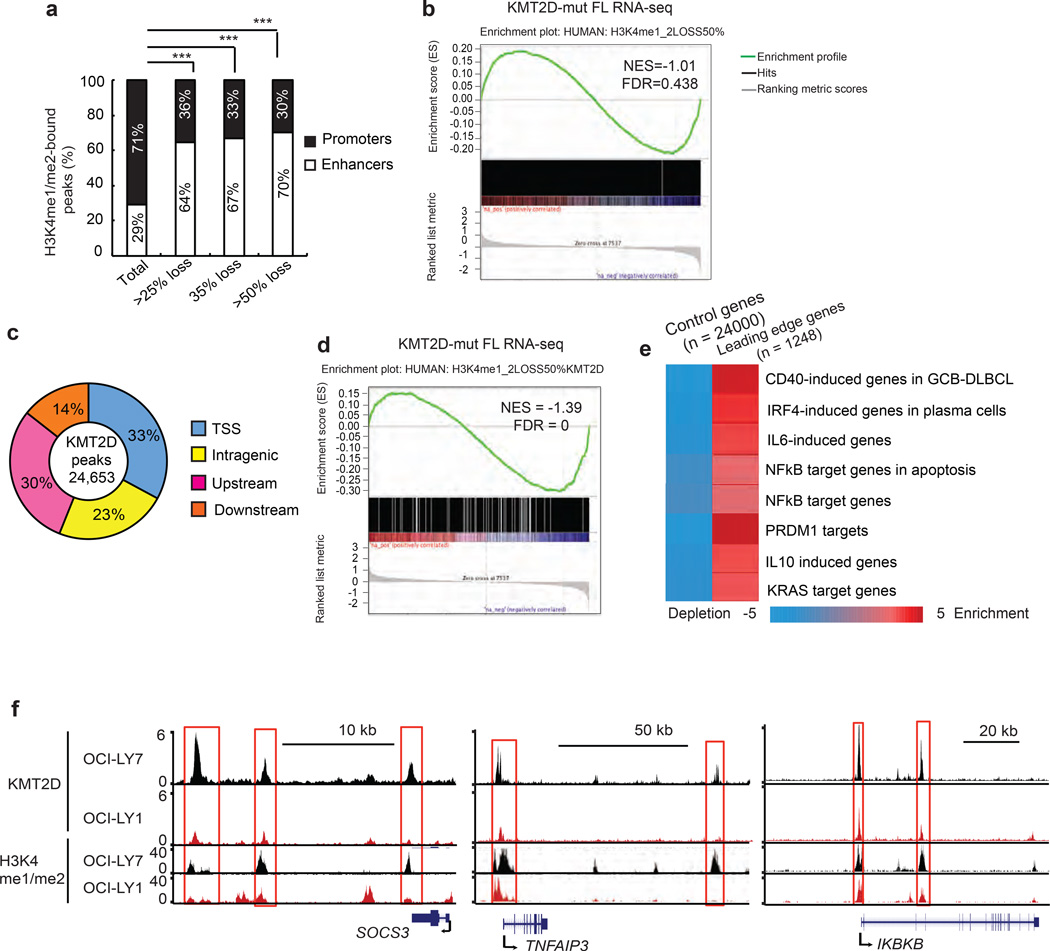
To assess how KMT2D depletion contributes to transcriptional regulation, we measured H3K4 mono- and di-methylation (H3K4me1/me2) in Kmt2d deficient and control lymphomas. We performed ChIP-seq using an antibody that specifically recognizes H3K4me1/me2 on DNA, from purified B220+ mouse lymphoma cells (n=3 for both VavP-Bcl2/vector and VavP-Bcl2/sh-Kmt2d). First, analysis of ChIP-seq data for H3K4me1/me2 abundance did not reveal a global loss of the marks genome wide (Fig. 4a). We confirmed this observation by immunoblots for H3K4me1/me2/me3 on lysates form sorted B220+ mouse Kmt2d knockdown lymphoma cells (Supplementary Figs. 4a, b) and non-malignant B220+ cells from wild type and Kmt2d−/− mice (Supplementary Figs. 4c, d). By contrast, we observed focal depletion of the H3K4me1/me2 marks at a subset of genomic sites in the mouse VavP-Bcl2/sh-Kmt2d lymphomas. Specifically, H3K4me1/me2 depletion was significantly more pronounced at putative enhancers compared to promoter elements (Fig. 4b, Supplementary Table 5). Using gene set enrichment analyses (GSEA) we found that the genes associated with significant H3K4me1/me2 depletion in enhancers and promoters (≥25% read density reduction) were significantly enriched among down regulated genes in both mouse and human KMT2D deficient lymphomas (Figs. 4c, d, Supplementary Figs. 4e, f). The leading edge genes (Supplementary Table 4) driving this association were significantly enriched for target genes induced by CD40, NFkB, IL6, IL10, LPS, TGFβ and TNF (Fig. 4e; Supplementary Fig. 4g; Supplementary Table 4). Notably, among the genes showing depletion of the H3K4me1/me2 mark at enhancers and concurrent changes in gene expression were tumor suppressor genes such as TNFAIP3 (A20)14, SOCS315, TNFRSF14 (HVEM)16, ASXL1, and ARID1A (Fig. 4f, Supplementary Fig. 4h).
Figure 4. Epigenetic effects of KMT2D on target genes in mouse lymphomas.
(a). Average H3K4me1/me2 read density plot at promoters and enhancers in MACS-purified B220+ positive B cells from VavPBcl2-vector and VavPBcl2-shKmt2d lymphomas identified by ChIP-seq. (b). Proportion of H3K4me1/me2 peaks by location near promoters or enhancers based on ChIP-seq on purified mouse B220 cells from VavPBcl2-vector and VavPBcl2-shKmt2d lymphomas. The proportion of affected promoters and enhancers for indicated thresholds (*** p < 0.001 by Chi-square). (c) and (d). GSEA of genes with a ≥25% reduction in H3K4me1/me2 marks at enhancers (p<0.05) in mouse B220 cells from VavPBcl2-vector and VavPBcl2-shKmt2d lymphomas compared to ranked gene expression changes (log2 fold) in B220+ mouse lymphomas (c), or in KMT2D wild type and mutant human FL specimens (d); NES, normalized enrichment score. FDR, false discovery rate. (all analyses: GSEA FDR<0.002) (e). Pathway analysis of down regulated genes (n=322) identified in the GSEA leading edge analyses in b and c compared to lymphoid signature database from the Staudt Lab (http://lymphochip.nih.gov/signaturedb/) and MysigDB. The background included around 24,000 genes from Refseq gene annotation. Statistical signficance was determined by hypergeometric test and shown in the color key. The red color indicates (in log10) the over-represented p-values and the blue shows under-representation. (f). Normalized UCSC read density tracks of H3K4me1/me2 ChIP-seq peaks from sh-Kmt2d (red) and vector (black) B220+ mouse lymphomas.
Next, we analyzed H3K4me1/me2 abundance in human lymphoma cells lines that were either KMT2D wild type (OCI-LY7, HT, DOHH2, SU-DHL4) or mutant (OCI-LY1, OCI-LY18, Toledo, Karpas422). Similar to the mouse lymphomas, measurements of global H3K4 methylation by immunoblotting and mass spectrometry showed no difference between KMT2D wild type and mutant lymphoma lines (Supplementary Figs. 5a–c). Consistent with the mouse lymphomas, H3K4me1/me2 ChIP-Seq on KMT2D wild type (OCI-LY7) and mutant (OCI-LY1) lymphoma cells showed a focal defect, limited to a subset of H3K4me1/me2 sites and ranking by the extent of H3K4me1/me2 depletion confirmed a predominant effect on enhancers similar to the experiments in the mouse lymphoma cells (Fig. 5a; Supplementary Table 5). However, initial analyses for enrichment of H3K4me1/me2 loss among genes that were down regulated in KMT2D mutant human FLs did not show the expected enrichment (Fig. 5b, Supplementary Table 6). This indicated that we needed to more accurately define the direct KMT2D target genes in human lymphoma cells. In order to measure directly KMT2D binding we performed ChIP-seq using a validated antibody against KMT2D in OCI-LY7 and OCI-LY1 cells. Similar to previous studies we identified ~ 24,000 binding sites; 32% were associated with transcriptional start sites (TSS) and the others were distributed to upstream, downstream, and intragenic locations in OCI-LY7 cells (Fig. 5c)5. Strikingly, genes that had a loss of H3K4me1/me2 with direct KMT2D binding in OCI-LY7 cells, were highly significantly enriched among the down regulated genes identified in FL subjects with KMT2D mutations (Fig. 5d; Supplementary Table 6). Once again these KMT2D target genes were significantly associated with immune signaling pathways including CD40, IL6, IL10, NFkB, IRF4 and others (Fig. 5e and Supplementary Table 4). Consistent with the analysis of the mouse lymphomas, these genes included the lymphoid tumor suppressors TNFAIP3 (A20) and SOCS3 with consistent changes in KMT2D binding and H3K4 methylation in KMT2D wild type (OCI-LY7) and mutant (OCI-LY1) cells (Fig. 5f; Supplementary Fig. 5d).
Figure 5. Identification of KMT2D target genes in human lymphoma cells.
(a). Proportion of H3K4me1/me2 peaks by location near promoters or enhancers by ChIP-seq on OCI-LY1 (KMT2Dmut) compared to OCI-LY7 (KMT2Dwt) cells for indicated thresholds (*** p < 0.001 by Chi-square). (b). GSEA of genes with a ≥50% reduction in H3K4me1/me2 read density in OCI-LY1 (KMT2Dmut) vs. OCI-LY7 (KMT2Dwt) cell lines compared to genes ranked by log2 fold change in KMT2Dwt vs. KMT2Dmut FL specimens. (c). Genomic distribution of KMT2D peaks located at transcript start sites (TSS), inside gene bodies (intragenic), or upstream and downstream of the closest gene in OCI-LY7 cells. (d). GSEA of genes with KMT2D binding in OCI-LY7 cells and >50% reduction in H3K4me1/me2 marks in OCI-LY1 (KMT2Dmut) vs. OCI-LY7 (KMT2Dwt) compared to genes ranked by log2 fold change in wild type and mutant human FL specimens (GSEA FDR=0). (e). Pathway analysis of down-regulated genes in FL subjects (p<0.05 by Wald test) with a KMT2D binding and >50% reduction in H3K4me1/me2 mark in OCI-LY1 vs, OCI-LY7 cells (n=1248) compared to lymphoid signature database from the Staudt Lab (http://lymphochip.nih.gov/signaturedb/) and MysigDB. The background included around 24,000 genes from Refseq gene annotation. Statistical signficance was determined by hypergeometric test and shown in the color key. The red color indicates (in log10) the over-represented p-values and the blue shows under-representation. (f). Normalized UCSC read density tracks of KMT2D ChIP-seq peaks in OCI-LY7 (black), OCI-LY1 (red).
Functional validation of selected KMT2D target genes
Based on concordant changes in expression, H3K4me1/me2 depletion, and KMT2D binding we selected several candidate KMT2D targets for further validation (SOCS3, TNFSRF14, TNFAIP3, ARID1A, DUSP1, TRAF3, NR4A1, IKBKB, DNMT3A, ASXL1, ARID3B, MAP3K8 and SGK1). First, we generated isogenic pairs of parental and KMT2D knockdown human lymphoma cells based on the KMT2D wild type lines (OCI-LY7 and SU-DHL4). Unlike in certain solid tumor cells17, KMT2D deficient lymphoma cells were significantly more proliferative in vitro than their parental counterparts (Supplementary Fig. 6a, b). Next, we tested three additional shRNAs for KMT2D knockdown (sh-KMT2D #1–3) and used qRT-PCR to measure effects on candidate target gene expression. Results showed a significant loss of expression for the indicated KMT2D targets in both isogenic, paired human cell lines and also in purified mouse lymphoma B220+ B cells (Fig. 6a, Supplementary Fig. 6c–e). For further confirmation of KMT2D mediated H3K4me1/me2 methylation at enhancer regions, we performed H3K4me1/me2 quantitative ChIP (qChIP) on the isogenic pairs of KMT2D knockdown and parental OCI-LY7 cells. We observed significant loss of H3K4me1/me2 in the enhancer regions of SOCS3, TNFAIP3, TRAF3, SGK1 and IKBKB, compared to TNS4, which is not a KMT2D target gene (Fig. 6b). Hence, KMT2D targets the regulatory regions of several tumor suppressor genes that control B cell signaling pathways.
Figure. 6. KMT2D inactivation affects growth and survival pathways in lymphoma cells.
(a). mRNA levels measured by qRT-PCR in the isogenic OCI-LY7 pairs expressing vector control or an shRNA against KMT2D. Values correspond to average of three replicates ± s.d., two-tailed Student’s t-test: *p<0.05, **p<0.01, ***p<0.001. (b). H3K4me1/me2 occupancy loss in enhancer regions of specified KMT2D target genes after KMT2D knockdown in OCI-LY7 cells by qChIP analysis. A genomic region (TNS4) with no KMT2D binding and H3K4me1/me2 was used as a negative control. Values correspond to mean percentage of input enrichment ± SD of triplicate q-PCR reactions of a single replicate. Two-tailed Student’s t-test was used to determine statistical significance (***p<0.001). Data correspond to one representative assay from a total of 2–3 independent assays. (c). Immunoblot of the indicated proteins in vector or sh-KMT2D expressing OCI-LY7 cells upon 48h of IL-21 stimulation. (d). TNFAIP3 mRNA levels in OCI-LY7 and SU-DHL4 transduced with vector or KMT2D-shRNA upon 48h anti-CD40 and IgM stimulation. Values correspond to average of three experimental replicates ± s.d., and statistical significance was determined by the two-tailed Student’s t-test: *p<0.05, **p<0.01, *p<0.001. (e). Flow cytometric analysis of cell death induced by anti-CD40 and anti-IgM treatment in OCI-LY7 lymphoma transduced with vector or KMT2D-shRNA. (f). Proliferation of OCI-LY7 (KMT2Dwt), and OCI-LY1 (KMT2Dmut) lymphoma cell lines upon stimulation with antiCD40 or anti-CD40 and anti-IgM. Values correspond to average of three experimental replicates relative to day 0± s.d. Two-tailed Student’s t-test was used to determine statistical significance. *p<0.05, **p<0.01. (g) and (h). Viability assay in lymphoma cell lines analyzed by flow cytometry using annexin-V and DAPI exclusion upon stimulation with anti-CD40 for 96h (g); quantification (h). Bars correspond to average of three experimental replicates ± s.d. Two-tailed Student’s t-test was used to determine statistical significance. *p<0.05, **p<0.01. (i) TNFAPI3/A20 mRNA levels (left) and NFKBIZ mRNA levels (right) in OCI-LY7, HT, SU-DHL4 (KMT2Dwt), OCI-LY1 and NU-DUL1 (KMT2Dmut) lymphoma cell lines upon anti-CD40 or anti-CD40 and IgM stimulation (24h). Bars represent mean of 3 biological replicates (2 biological replicates for NU-DUL1 anti-CD40+IgM) ± s.d. Two-tailed Student’s t-test was used to determine statistical significance *p<0.05, **p<0.01. Red labels represent KMT2Dmut cell lines and black labels represent KMT2Dwt cell lines.
Next, we probed how KMT2D loss in human lymphoma cells affected the specific functions of key KMT2D targets. We generated isogenic pairs of KMT2D proficient and deficient human lymphoma cell lines using shRNA knockdown. For example, we identified SOCS3, a negative regulator of STAT3 signaling, as a KMT2D target. Accordingly, we find a reduction of SOCS3 protein and an augmentation in the JAK-STAT response upon IL21 stimulation in the KMT2D deficient cells compared to isogenic control OCI-LY7 cells (Fig. 6c). We also identified key signaling molecules related to the CD40, B cell receptor (BCR), and Toll-like receptor (TLR) pathways among the KMT2D target genes (e.g. TRAF3, TNFAIP3, MAPK3K8, DUSP1). Transcriptional expression of many of these target genes in B cells is dependent on CD40 and BCR signal activation, including TNFAIP318,19. Therefore, we tested if KMT2D loss in KMT2D WT cell lines, OCI-LY7 and SU-DHL4, affected the induction of KMT2D target genes by anti-CD40 and anti-IgM. Analysis by qRT-PCR showed that the induction of TNFAIP3 upon anti-CD40 and anti-IgM treatment was abrogated in both cell lines after KMT2D knockdown (Fig. 6d). CD40 signaling has also been shown to be pro-apoptotic in a panel of DLBCL cell lines20. Therefore we tested if KMT2D knockdown could protect OCI-LY7 cells from anti-CD40 induced apoptosis. OCI-LY7 cells harboring the KMT2D shRNA showed reduced cell death induction by annexin V and DAPI staining after anti-CD40 and IgM treatment (Fig. 6e). We made analogous observations when comparing panels of KMT2D wild type (OCI-LY7, HT, SU-DHL4) and KMT2D mutant (OCI-LY1, OCI-LY18, NU-DUL1) lymphoma cell lines. For example, the KMT2D wild type OCI-LY7, HT and SU-DHL4 cells show a greater growth inhibition compared to the KMT2D mutant cell lines OCI-LY1, OCI-LY18 and NU-DUL1 upon treatment with anti-CD40 alone or in combination with anti-IgM (Fig. 6f, Supplementary Fig. 6g). Similarly, viability assays showed that KMT2D wild type cells were more sensitive to anti-CD40 treatment and had increased levels of apoptosis, measured by annexin V and DAPI staining, compared to mutant cells (Fig. 6g, h). These differences were not caused by differential CD40 receptor expression, as only OCI-LY18 did not express CD40 receptor and was not affected by anti-CD40 treatment (Supplementary Fig. 6f). Analysis of target gene expression showed an overall attenuated transcriptional response in KMT2D mutant cell lines for important KMT2D targets such as the tumor suppressors genes TNFAIP3/A20, NFKBIZ, FAS, and DUSP1 (Fig. 6i, Supplementary Fig. 6h). Hence, KMT2D deficiency affects key effects of BCR, CD40, and JAK-STAT signaling in lymphoma B cells.
Discussion
Our results establish the tumor suppressor function of KMT2D in germinal center B cells. The H3K4 methyltransferase KMT2D is one of the most frequently mutated genes in DLBCL and FL3,4 and we show that it controls the expression of multiple key regulators of the CD40, Toll-like, and B cell receptor signaling pathways (Supplementary Fig. 7). Bona fide KMT2D target genes include lymphoid tumor suppressor genes such as TNFAIP3, SOCS3, SGK1, TRAF3, TNFRSF14, and ARID1A15,16,21. KMT2D also contributes to the normal B cell response and KMT2D deficient mice show an abnormal persistence of germinal centers, a defect in class switch recombination, and reduced antibody production reminiscent of the reported immune defect seen in the heritable Kabuki syndrome that is most often linked to KMT2D mutations. Collectively these data suggest that KMT2D somatic mutation may drive GC expansion due to enhanced proliferation and impaired terminal differentiation, due to loss of H3K4 mono and di-methylation at key B-cell enhancer regions and some promoters. Our results are consistent with genomic evidence indicating that KMT2D mutations are early lesions in GC lymphomas3,4. Notably, even in the absence of Bcl2 activation, KMT2D deficiency is sufficient to trigger B cell malignancy in mice. Clinically, KMT2D mutations are not associated with the outcome of R-CHOP chemotherapy in DLBCL. However it is not yet known how KMT2D status would affect the response to targeted signal inhibitors that are entering the clinic. In this regard, our results indicate deregulation of multiple immune signaling pathways in KMT2D mutant lymphoma cells and altered responses to anti-CD40 and B-cell receptor activation. Recently, histone deacetylase (HDAC) inhibitors were shown to ameliorate the developmental defects in a model of Kabuki syndrome22. Similarly, inhibition of H3K4 demethylase activities such as JARID1 and LSD1 may be able to reverse some of the epigenetic changes seen in KMT2D deficient lymphomas23.
Online Methods
Measurement of KMT2D mRNA expression in human B cells
The human tonsil and bone marrow samples were obtained in Pamplona (Spain), Clinica Universidad de Navarra and the obtention of these samples were approved from the ethical committee of Clinica Universidad de Navarra (Spain). Cells from tonsils/bone were immunophenotyped using a single 8-color antibody combination [Pacific Blue (PB)/ Oranje Chrome 515 (OC515)/ fluorescein isothiocyanate (FITC)/ phycoerythrin (PE)/ peridinin chlorophyll protein-cyanin 5.5 (PerCP-Cy5.5)/ PE-cyanin 7 (PE-Cy7)/ allophycocyanin (APC)/ APCH7] - CD20/ CD45/ CD38/ CXCR4/ CD3/ CD10/ CD27/ CD44 - aimed at the identification and high-purity (≥97%) FACS-sorting (FACSAria II, Becton Dickinson Biosciences, San Jose, CA) of the following B cell (CD3− / CD20+ / CD45+) subsets (after careful exclusion of CD3+ / CD20−/ CD45+ T-cells): naïve B cells (CD10−/ CD44+/ CD27−/ CD38−), germinal center (CD10+/ CD44lo/ CD38+) centrocytes (CXCR4−) and centroblasts (CXCR4+), memory B cells (CD10−/ CD27+/ CD44+) and newborn plasmablasts (CD10−/ CD27hi/ CD38hi/ CD44hi). The strand specific RNA-seq was performed in naïve B cells (n=5 samples), centroblasts (n=7), centrocytes (n=7), memory cells (n=8), tonsilar plasma B cells (n=5) and purified plasma B cells from bone marrow of healthy donors (n=3). Each red dot represents a separate human tonsil and the mean expression is represented in TPM (transcripts per million).
Characterization of human FL samples
Institutional Review Board of Weill Cornell Medical College (IRB#0107004999) approved the study protocol. The specimens are derived from excess diagnostic materials that were banked in the lymphoma repository. A waiver of informed consent has been obtained for this retrospective study. The IRB- approved protocol permits association of these specimens with a particular individual, allowing review of the medical records for the minimum information necessary to complete the study. All data provided to investigators are stripped of protected health information.
Sample preparation
Frozen single cell suspension of individual tumor sample was first thawed in a 37°C water bath, and then resuspended in RPMI + 10%FBS and incubated in a incubator (37°C and 5% CO2) for 1 hr. Half of the sample was used to isolate B cells by using EasySep Human B Cell Enrichment Kit (STEMCELL Technologies, Vancouver, Canada), and the other half was used to isolate T-cells with Easy Sep Human T Cell Isolation Kit. DNA was extracted from isolated cell populations by using PureLink Genomic DNA kit (LifeTechnologies, Grand Island, NY). Total RNA was extracted by using Qiagen RNeasy Mini Kit (Valencia, CA). The quantity of DNA and RNA samples was measured by a Qubit Fluorometer (LifeTechnologies, Grand Island, NY), and the quality of DNA and RNA samples was assessed by a bioanalyzer (Agilent Technologies, Santa Clara, CA).
Exome sequencing
For each tumor sample and the respective T-cell control sample, 3 µg high molecular weight genomic DNA was used to prepare exome sequencing libraries using the Aglient SureSelectXT Human All Exon 50 MB Target Enrichment System for Illumina Pair-End Sequencing Library kit (Agilent Technologies, Santa Clara, CA). Each library was sequenced on one entire lane of flowcell on Illumina HiSeq 2000. Sequence information of 75 bp on each end of the DNA library fragment (PE75) was collected.
Targeted re-sequencing
A targeted-enrichment panel was designed by RainDance Technologies (Billerica, MA) for 36 most commonly mutated lymphoma genes including, ARID1A, ATP6AP1, B2M, BCL2, BCL6, BTG1, BTG2, CARD11, CD79B, CREBBP, EB1, EEF1A1, EP300, EZH2, GNA13, HIST1H1B, HIST1H1C, HVCN1, IRF4, IRF8, KLHL6, KMT2D, MEF2B, MYD88, PCGF5, PDS5A, PIM1, POU2F2, PRDM1, SGK1, STAT6, SZT2, TBL1XR1, TNFAIP3, TP53, and XPOT. The entire coding regions of this set of genes were targeted by overlapping PCR amplicons averaging 200 bp. DNA (200 ng) was first sheared to around 3 kb by using a Covaris S220 Focused-ultrasonicator (Woburn, MA), then merged with primer pairs in picoliter-droplets format on a Raindance ThunderStorm system. Targeted regions were amplified with the addition of specific tailed primers. A second round of PCR was performed to add indexed adaptor sequences for Illumina sequencing. Final indexed products from 48 samples were multiplexed together and sequenced on one entire lane of flowcell on Illumina HiSeq 2500 by using the fast mode setting. Sequence information of 100 bp on each end of the library fragment (PE100) was collected.
SNV discovery
Sequencing reads were aligned to human genome assembly GRCh37/hg19 using the BWA aligner24. After filtering duplicated paired reads, variants were detected as previously described25–27. Novel coding region SNVs were defined as those were not present in SNP132. These SNVs were then further filtered by sequencing depth (≥20×) and variant percentage (≥25%). To obtain the list of somatic mutations in each tumor sample, we compared the variant ratio of each novel coding SNV between tumor B cells and respective control T-cells, and estimated the statistical significance of the difference by using a Chi-square tested corrected with multiple hypothesis testing (Benjamini-Hochberg corrected P < 0.1).
Characterization of DLBCL samples
We analyzed 347 newly diagnosed DLBCL cases that were treated with R-CHOP given with curative intent at the BC Cancer Agency (Vancouver). Subject sample use was approved by the University of British Columbia, British Columbia Cancer Agency, Research Ethics Board (REB #H13-01478). The cases were selected on the basis of the following criteria: 16 years of age or older; histologically confirmed de novo DLBCL according to the 2008 WHO classification; available DNA extracted from fresh frozen biopsy material (tumor content >30%). All cases were centrally classified by AM and RDG who were blinded for sample identity to determine the diagnosis. Individuals were excluded if they were younger than 16 years old and had other than de novo DLBCL (primary mediastinal large B cell lymphoma, primary central nervous system lymphoma and a previous diagnosis of an indolent lymphoproliferative disorder) and positive HIV serology.
Targeted resequencing in DLBCL samples
Targeted re-sequencing of the coding exons of KMT2D in 347 DLBCL cases was performed using a Truseq Custom Amplicon assay (Illumina) and libraries were run on the Miseq. Mutation calling was done with Mutascope pipeline. COO classification was available in 331 cases, according to gene expression profiling by the Lymph2Cx assay using the NanoString platform28 in 299 subjects as well as Hans algorithm29 in 32 cases with low tumor content. 194 cases were assigned to GCB subtype, 107 cases, ABC/non-GCB and 30 were unclassifiable.
Correlation between KMT2D mutation status with disease progression and survival
Baseline characteristics were compared between the groups with KMT2D mutation type using chi-square test.
We measured the endpoints from the time of the initial pathologic diagnosis to the following events; overall survival (OS; the date of death from any cause or to the last follow-up); progression-free survival (PFS; the date of progression, relapse or death from any cause); disease-specific survival (DSS; the date from lymphoma or acute treatment toxicity) and time-to-progression (TTP; the date of progression, relapse or death from lymphoma or acute treatment toxicity). OS, PFS, DSS and TTP were estimated using the Kaplan-Meier method and differences in outcome between groups were assessed using the log-rank test. Two-sided P<0.05 was considered significant. Data were analyzed using SPSS software (SPSS version 14.0; SPSS Inc, IL).
Generation of mice
Kmt2df/f mice were previously described7 and here we bred them with CD19-Cre mice (Jackson no. 006785) where Cre is expressed from the pre-B cell stage and removes exons 16–19 of Kmt2d causing an open reading frame shift that creates a stop codon in exon 20. Kmt2df/f CD19-Cre mice were maintained in a mixed C57BL/6; 129 background. Mice were monitored for tumor formation once a week for the first 4 months and every day after then. All mice were housed in the Frederick National Laboratory and treated with procedures approved by the NIH Animal Care and Use Committee.
The vavP-Bcl2 mouse model of FL9 was adapted to the adoptive transfer approach using retrovirally transduced HPCs. HPCs isolation and transduction were performed as in30. 8–10 week old C57BL/6 females lethally irradiated (4.5Gy twice) were used as recipients for all transplantation experiments. Mouse Kmt2d shRNAs were design using Designer of Small Interfering RNA (DSIR, http://biodev.extra.cea.fr/DSIR/) and are based on MSCV31
sh-Kmt2d #1 (mouse): GACTGGTCTAGCCGATGTAAA.
sh-Kmt2d #2 (mouse): TGAATCTTTATCTTCAGCAGG
Mouse B220+ tumor sample preparation
B220+ cells were purified from mouse lymphoma tumors by immunomagnetic enrichment with CD45R(B220) microbeads (Miltenyi Biotech). RNA extraction was performed using TRIzol (Ambion) following the manufacturer’s protocol.
Histology
Mouse tissues were fixed overnight in formalin, embedded in paraffin blocks and sectioned. Tissue sections were stained with hematoxilin/eosin (H&E) or with Ki67, Tunel, B220, and PNA following standard procedures32,33
Flow cytometric analysis
VavP-Bcl2 tumors
Tumor cell suspensions of representative tumors of each genotype were stained as described30. The antibodies used were B220/CD45R (BD PharMingen, #553092), IgG1 (BD PharMingen #560089) conjugated with APC, B220/CD45R (BD PharMingen, #553090), CD19 (BD PharMingen, # 557399), IgM (PharMingen, #553409), Thy1/CD90 (Cedarlane, #CL8610PE), CD8 (PharMingen, #553032), Sca-1 (PharMingen, #553108), IgD (BD PharMingen #558597), GL7 (BD PharMingen #561530) conjugated with phycoerythrin and analyzed with BD LSRFortessa™ cell analyzer. Analysis was performed with FlowJo software (Tree Star).
Kmt2d−/− tumors
Single-cell suspensions were obtained from spleens according to standard procedures. Red blood cells were lysed with ACK solution and surface markers on tumor cells were analyzed on FACSCalibur (BD Biosciences) using the following fluorochrome-cojugated antibodies: IgM-PE (BD Pharmingen, clone R6-60.2 #553409), IgM-FITC (BD Pharmingen, clone R6-60.2 #553408), IgD-FITC (BD Pharmingen, clone 11-26c.2a #553439), Igλ1, λ2, λ3 -FITC (BD Pharmingen, clone R26-46 #553434), Igκ-FITC (BD Pharmingen, clone 187.1 #550003), CD19-APC (BD Pharmingen, clone 1D3 #550992), B220-PE (BD Pharmingen, clone RA3-6B2 #553090), B220-PE (BD Pharmingen, clone RA3-6B2 #553088), CD138-PE (BD Pharmingen, clone 281.2 #553714), CD24-FITC (BD Pharmingen, clone M1/69 #553261), CD11b-APC (BD Pharmingen, clone M1/70 #553312), CD4-PE (Biolengend, clone GK1.5 #100408), CD8-FITC (BD Pharmingen, clone 53-6.7 #553031), CD3-PE (BD Pharmingen, clone 500A2 #553240) and CD43-biotin (BD Pharmingen, clone S7 #553269) and B220-biotin (BD Pharmingen RA3-6B2 #553085) followed by Streptavidin-APC (BD Pharmingen). Analysis was performed with FlowJo software (Tree Star).
Characterization of nonmalignant B cell populations in Kmt2d−/− mice
To identify the different B cell populations, two stains were performed in splenocytes from 4 to 5.5 month old mice (2 females and 1 male wt, and 4 females Kmt2d−/−). First, to identify transitional, follicular and marginal zone populations, cells were stained with CD21-FITC (Biolegend, clone 7E9, #123407), CD5-PE (eBioscience, clone 53–7.3 #12-0051-81), CD23-PECY7 (Biolegend, clone B3B4 #101613), IgM-APC (Biolegend, clone RMM-1 #406509), and B220-Alexa700 (Biolegend, clone RA3 #103232). To identify intermediate plasma cells/plasmablast (IPC), plasma cells (PC) and germinal center populations, cells were stained with GL7-FITC (Biolegend, clone GL7 #144003), CD138-PE (Biolegend, clone 281-2 #142503), CD95-APC (eBioscience, clone 15A7 #17-0951-80), B220-Alexa700 (Biolegend, clone RA3 #103232). To determine the percentages of cell populations, values were normalized by % B220+ single live cells (single cells, 7-AAD negative. B220+). 7-AAD (Life Technologies) was used to identify dead cells. Data acquisition was performed in a BD LSR II Flow Cytometer (BD Biosciences) and analysis was performed with FlowJo software (Tree Star).
DLBCL cell lines
CD40r expression on DLBCL cell lines was measured using FITC conjugated anti-CD40 (BD clone C53 #B555588). DLBCL cell line viability was measured by APC conjugated anti-annexinV (BD #B550474) and DAPI exclusion. Data were acquired on MacsQuant flow cytometer (Miltenyi Biotec) and analyzed using FlowJo software package (TreeStar).
IgVH rearrangement analysis
PCR to evaluate IgVH rearrangements was performed on cDNA of VavP-Bcl2 lymphoma cells with a set of forward primer that anneal to the framework region of the most abundantly used IgVL gene families and reverse primer located in the Jλ1,3 gene segment (IgL- Vλ1: GCCATTTCCCCAGGCTGTTGTGACTCAGG and IgL-Jλ1,3: ACTCACCTAGGACAGTCAGCTTGGTTCC)34
Class Switch Recombination (CSR) in Kmt2d−/− tumors
Genomic DNA isolated from tumors cell suspensions and MEFS as a germinal band control were restricted and for southern blot hybridization was performed with the following probes: JH probe (PCR amplified with 5'-TATGGACTACTGGGGTCAAGGAAC-3' and 5'-CCAACTACAGCCCCAACTATCCC-3', personal communication), 3'Smu probe (PCR amplified with 5'-CCATGGGCTGCCTAGCCCGGGACTTCCTGCCC and 5'-ATCTGTGGTGAAGCCAGATTCCACGAGCTTCCCATCC-3',), and IgκIII (an EcoRI/SacI fragment of Igκ locus, gift from Dr. Barry Sleckman)
Somatic Hypermutation
The genomic sequences from VH to the intron downstream of JH4 were PCR-amplified from tumor DNA using degenerate forward primers for the different VH families35 and a reverse primer (5'-AGGCTCTGAGATCCCTAGACAG-3')36 downstream JH4. Proofreading polymerase (Phusion High Fidelity, NEB) was used for amplification with PCR conditions previously published35. Amplification products were isolated from agarose gel and submitted to sanger sequencing. Sequences were compared with reference and mutation rate calculated using IMGT/V-QUEST37 and UCSC BLAT. PCR amplification and sequencing was repeated two or three times for each sample. As a negative and positive control DNA extracted from MEFS and Igκ-AID B cells respectively were used in parallel (not shown).
Characterization of mouse B cell differentiation and antibody production
Germinal center assessment in mice
HPCs from C57BL/6 mice were retrovirally transduced with empty vector or sh-Kmt2d and adoptive transfer approach was performed in 2 month old C57BL/6 females irradiated with 4.5Gy (n=3–4 per group). After four and seven weeks after injection of HPCs, females were immunized intraperitoneally with 0.5ml of 2% sheep red blood cell (SRBC) suspension in PBS (Cocalico Biologicals). Nine weeks later spleens were collected for histology and immunohistochemistry analysis. Ki67 positive cells were quantified using Metamorph software.
For analysis of the formation of GCs in Km2d−/− mice, 4 mice for each genotype (1.5–2 month old, wt: 2 males and 2 females; Kmt2d−/− mice: 3 males and 1 female) were immunized intraperitoneally with 100 µg NP21-CGG (Biosearch Technologies) in Imject alum (Pierce). On day 6 after immunization, splenocytes were harvested and B cell populations were analyzed by flow cytometry as above (see Characterization of B cell populations in Kmt2d−/− mice).
ELISA analysis of NP specific antibody production
Serum from NP-CGG immunized Kmt2d+/+ (wt) or Kmt2d−/− mice was analyzed for NP-specific IgM or IgG1 titer using the SBA Clonotyping System-HRP (SouthernBiotech). Plates were coated with 10ug/mL NP(20)-BSA (Biosearch Technologies) and serum from immunized or non-immunized mice were added to 96 well assay plates (Costar) at increasing dilutions in PBS with 1% BSA. Bound antibodies were detected with HRP-labeled goat anti-mouse IgG1 or IgM antibodies. The optical density of each well was measured at 405nm.
In vitro class switch recombination
For class switch recombination to IgG1 resting splenic B cells were isolated from 2.5 to 5 months old Kmt2d+/+ CD19-Cre- (wt, 2 females and 3 males) and Kmt2df/f CD19-Cre+ (Kmt2d−/− 2 females and 3 males) mice by immunomagnetic depletion with anti-CD43 MicroBeads (anti-Ly48, Miltenyi Biotech), and cultured at 0.5×106 cells/ml with LPS (25 µg/ml; Sigma), IL-4 (5 ng/ml; Sigma) and RP105 (Anti-Mouse CD180; 0.5 µg/ml; BD Pharmingen) for 4 days. B cells were infected at 24 and 48 hours in culture with pMX-Cre-IRES-GFP as described38 to enhance Kmt2df/f deletion. Class switching to IgG1 was measured at 96 hours in the GFP+ population (>90%) by flow cytometry using the following antibodies: IgG1-biotin (BD Pharmingen, clone A85-1 #553441) following streptavidin-Pacific Bule (Molecular Probes), B220-Alexa700 (Biolegend, clone RA3 #103232). Data acquisition was performed on the BD LSR II Flow Cytometer (BD Biosciences) equipped with CellQuest software (Becton Dickinson). Analysis was performed with FlowJo software (Tree Star).
mRNA-seq Library Preparation and Sequencing analysis
RNA was purified using the RNAeasy Plus Kit (QIAGEN) that included a genomic DNA elimination step. RNA size, concentration, and integrity was verified using Agilent 2100 Bioanalyzer (Agilent Technologies). Libraries were generated using Illumina’s TruSeq RNA sample Prep Kit v2, following the manufacurer’s protocol. Sequencing of 8–10 pM of each library was done on the HiSeq2500 sequencer as 50bp single read runs. RNA-seq data from mouse B220 cells were aligned to the mm9 genome using STAR. RNA-seq data from FL subjects were aligned to the hg19 genome using TopHat. −2.0.10 with default parameters except −r 150 [TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions]. Read counts were derived from HTSeq.scripts.count module in HTSeq-0.6.0 with default parameters [HTSeq—a Python framework to work with high-throughput sequencing data]. Differentially expressed genes were generated by DESeq2-1.6.3 in R [Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2].
ChIP and ChIP-seq Library Preparation and Sequencing analysis
H3K4me1/me2 ChIP was performed as previously described39. Briefly, 4×106 mouse B220+ cells or DLBCL cells were fixed with 1% formaldehyde, lysed, and sonicated (Branson Sonicator; Branson) leading to a DNA average size of 200 bp. 4ul of H3K4me1/me2 antibody (Abcam 32356 lot GR106705-5), tested for specificity by histone peptide array (Active Motif 13001), was added to the pre-cleared sample and incubated overnight at 4°C. The complexes were purified using protein-A beads (Roche) followed by elution from the beads and reverse crosslinking. DNA was purified using PCR purification columns (QIAGEN).
H3K4me1/me2 ChIP-seq libraries were prepared using 10ng of DNA and Illumina’s TruSeq ChIP sample prep, according to the manufacturer. Libraries were validated using the Agilent Technologies 2100 Bioanalyzer and Quant-iT dsDNA HS Assay (Life Technologies) and 8–10 pM sequenced on HiSeq2500 sequencer as 50bp single read runs. ChIP-seq data was aligned to the hg18 and hg19 genomes using STAR. Peak calling and read density in peak regions were performed by ChIPseeqer-2.1 with default parameters [An integrated ChIP-seq analysis platform with customizable workflows].
KMT2D ChIP assays were performed as previously described40. Briefly, 3–5×107 cells were crosslinked with 1% paraformaldehyde at room temperature for 15 min and sonicated to generate chromatin fragments of 200–600bp. Fragmented chromatin was then immunoprecipitated overnight with in-house generated human KMT2D antibody specific for the N-terminus previously described5, followed by washes and elution. ChIP-sequencing libraries were prepared with KAPA HTP ChIP-seq sample prep kit (KAPA Bioystems) for further high throughput sequencing.
H3K4me1/me2 ChIP DNA from KMT2D shRNA or empty vector control lentiviral transduced OCI-LY7 cells were quantified by qPCR. Primers were designed to amplify loci with KMT2D peaks in OCI-LY7 and H3K4me1/me2 depletion in OCI-LY1. Enrichment was calculated relative to input. The primers used were:
-
-TNFAIP3 (A20)
- Forward: GTGCTGCCATCCCCCAAATA,
- Reverse: AGCTTTCCCATGAGCCACT;
-
-SOCS3
- Forward: ACCTGGCTAGACTGAGGTCAT,
- Reverse: TTAGAGGCGCTCTGGTTCCT;
-
-TRAF3
- Forward: TCCAAGGGAAGATGAGGCCA,
- Reverse: CCTCGGGGGCCATAATACAG;
-
-SGK1
- Forward: GACCGATTGGGAAAGCAGGT,
- Reverse: GAGTTGGCTCTGGCTTCCAT;
-
-IKBKB
- Forward: AGGTCAACAAGGAGTCAGCC,
- Reverse: AGGAGGGAGGGGAGCTTTAT
-
-TNS4 (negative control loci)
- Forward: TTATTTGGCTGGGTGTGGT,
- Reverse: GTAGAGACGGGATTTCACCATG
Human_Downregulated_Genes are down-regulated genes (log fold change (logFC) < 0, p-val < 0.05, n=519) in FL subjects with nonsense KMT2D mutation versus WT KMT2D, Fig. 3e) based on RNA-seq data. Mouse_Downregulated_Genes were down-regulated genes (logFC < 0, p-adjust < 0.1, Benjamini–Hochberg method, n=1016) in mouse B220+ cells, sh-Kmt2d versus empty vector, Fig. 3f) based on RNA-seq data. We also determined an RNA-seq leading edge gene set (n=347, Fig. 3i). This gene set is the union of two sub- gene sets:- Top 200 down-regulated genes in Human_Downregulated_Genes (ranked by logFC derived from B220 RNA-seq)
- Top 200 down-regulated genes in Mouse_Downregulated_Genes (ranked by logFC derived from FL RNA-seq)
For H3K4me1/me2 ChIP data from mouse B220+ cells, candidate peaks were the union of the peaks called from each control replicate (n=3) with ChIPseeqer. We defined peaks overlapped with promoters (defined as ± 2kb windows centered on RefSeq transcription start sites (TSS)). Peaks that didn’t overlap with promoters, gene body and exons were treated as enhancer peaks. Enhancer peaks inside gene bodies were identified as intragenic enhancer peaks. Intergenic enhancer peaks were defined as being within a 50kb window from the corresponding genes. TSS Mouse_Enh_H3K4me1/me2_Loss were genes identified with H3K4me1/me2 depletion (>25% read density loss and p-val < 0.05, t-test, n=680) at enhancer peaks in sh-Kmt2d (n=3) (Figs. 4c, d). We also determined a Mouse H3K4me1/me2 ChIP-seq Enhancer leading edge gene set (n=322, Fig. 4e), which is the union of two sub-gene sets:- Top 200 down-regulated genes in Mouse_Enh_H3K4me1/me2_Loss gene set (ranked by logFC derived from B220 RNA-seq)
- Top 200 down-regulated genes in Mouse_Enh_H3K4me1/me2_Loss gene set (ranked by logFC derived from FL RNA-seq).
We derived Mouse_Pro_H3K4me1/me2_Loss gene sets (n=602, Supplementary Figs. 4e, f) and Mouse H3K4me1/me2 ChIP-seq Promoter leading edge genes (n=321, Supplementary Fig. 4g) in the same way as enhancers described above.
For H3K4me1/me2 ChIP data from OCI-LY1 and OCI-LY7 cell lines, candidate peaks were the union of the peaks called from two OCI-LY7 replicates (KMT2D WT) with ChIPseeqer. Promoter and enhancer peaks were determined by the same method described above for mouse B220 H3K4me1/me2 ChIPseq, In addition, all enhancer peaks were overlapped with annotated enhancers previously determined in OCI-LY7. Human_H3K4me1/me2_LOSS50 were genes with H3K4me1/me2 depletion (>50% read density loss, n=4416) in OCI-LY1 vs OCI-LY7 (Fig. 5b).
KMT2D peaks from KMT2D ChIP-seq data were called using ChIPseeqer. Human_H3K4me1/me2_Loss50_KMT2D were genes with H3K4me1/me2 loss peaks (>50% read density loss and overlapped with KMT2D peaks, n=1248, Fig. 5d). We chose 1248 genes as leading edge genes (ranked by H3K4me1/me2 loss from OCI-LY1 and OCI-LY7 ChIP-seq).
The code for performing these analyses will be made available upon request.
GO analysis with iPAGE
The GO analyses were performed with iPAGE41 The concept of mutual information (MI)42 to directly quantify the dependency between expression and known pathways MsigDB43 or in the lymphoid signature database from the Staudt Lab44 are used in iPAGE. Nonparametric statistical tests are then used to determine whether a pathway is significantly informative about the observed expression measurements. An iPAGE input file is defined across around 24,000 genes from Refseq genes, where each gene is associated with a unique expression status in our analysis. Meanwhile, each gene can be associated with a subset of M known pathways (e.g. from the Gene Ontology annotations). For each pathway, the pathway profile is defined as binary vector with N elements, one for each gene. “1” indicates that the gene belongs to the pathway and “0” indicates that it does not.
Given a pathway profile and a expression file with Ne groups, iPAGE creates a table C of dimensions 2×Ne, in which C(1, j) represents the number of genes that are contained in the jth expression group and are also present in the given pathway. C(2,j) contains the number of genes that are in the jth expression group but not assigned to the pathway. Given this table, we calculate the empirical mutual information (MI) as follows:
where
To assess the statistical significance of the calculated MI values, we used a non-parametric randomized-based statistical test. Given I as the real MI value and keeping the pathway profile unaltered, the expression file is shuffled 10,000 times and the corresponding MI values Irandom are calculated. A pathway is accepted only if I is larger than (1-max_p) of the Irandom values (max_p is set to 0.005). This corresponds to a p-value < 0.005. In iPAGE, pathways are first sorted by information (from informative to non-informative). Starting from the most informative pathways, the statistical test described above is applied to each pathway, and pathways that pass the test are returned. When 20 contiguous pathways in the sorted list do not pass the test, the procedure is stopped.
Highly statistically significant mutual information is explained by combination of over-representation and under-representation in specific expression groups. To quantify the level of over- and under-representation, the hypergeometric distribution is used to calculate two distinct p-values:
- For over-representation:
- For under-representation:
where x equals the number of genes in the given expression group which are also assigned to the give pathway. m is the number of genes assigned to the pathway (foreground), n is the number of genes in the expression group and N is the total number of genes (background). If pover < punder, we consider the pathway to be over-represented in the expression cluster; otherwise, it is under-represented. In the heatmap, the red color indicates (in log10) the over-represented p-values and the blue shows under-representation.
GSEA analysis
All the GSEA analysis results in this manuscript were generated from GSEA Pre-ranked mode43,45. There were two kinds of input files:
FL subjects: gene expression level logFC (Nonsense KMT2D mutation versus WT)
B220: gene expression level logFC (sh-KMT2D versus MLS)
In those input files, we chose the minimum logFC when a gene had multi-transcripts. All the gene sets used in GSEA were described in Computational Methods section.
Human cell lines
Lymphoma cell lines HT, DoHH2, SU-DHL4, Toledo, Karpas-442, OCI-LY8, NU-DUL1, and SU-DHL10 were maintained in RPMI 1640 with 10% fetal bovine serum, 1% L-Glutamine and 1% penicillin/streptomycin. OCI-LY7, OCI-LY1, and OCI-LY18 cells were cultured with IMDM media with 15% fetal bovine serum, 1% L-Glutamine and 1% penicillin/streptomycin. When indicated OCI-LY7 or SU-DHL4 lymphoma cells were transduced with lentiviruses expressing empty vector (pLKO.1) or shRNA against KMT2D (pLKO.1; Sigma, sh-KMT2D #1: TRCN0000013140; sh-KMT2D #2: TRCN0000013142; sh-KMT2D #3: TRCN0000235742). Source of cell lines are as follows, OCI-LY7=OCI, OCI-LY1=OCI, OCI-LY18=OCI, HT=ATCC, SU-DHL4=DSMZ, NU-DUL1=DSMZ. Cell lines were authenticated by STR DNA profiling by biosynthesis, (http://www.biosyn.com/celllinetesting.aspx). Mycoplasma contamination is routinely tested with Universal Mycoplasma detection kit ATCC (http://www.atcc.org/products/all/30-1012K.aspx).
Proliferation assays in lentiviral transduced OCI-LY7 cells were performed using Viacount assay fromGuava Technologies performed as reported46. 5×105 cells were seeded in in 2 ml into a single well of a 6 well dish. Each experiment was done in triplicate.
For IL-21 stimulation assay, lentiviral transduced OCI-LY7 cells with vector or shRNA against KMT2D were seeded and recombinant human IL-21 (PeproTech #200-21) was added to 10ng/ml final concentration; cells were collected after 48h and whole cell lysates were prepared.
For CD40/IgM stimulation assays, DLBCL cells were seeded at 2.5×105 cells in 500ul into a single well of a 12-well plate and cultured with anti-CD40 (2.5 ug/ml; RD Systems #AF632) alone or in combination with anti-IgM (10ug/ml; Jackson ImmunoResearch #109-006-129) for 1, 2 or 4 days. After 1 or 2 days, cells were collected for RNA isolation. After 4 days, cell death was measured using Annexin-V/DAPI staining.
Histone extraction and quantitative mass spectrometry analysis
Nuclei were isolated and histone proteins were extracted as described previously with minor modifications47. Briefly, histones were acid-extracted from nuclei with 0.2 M H2SO4 for 2 hours and precipitated with 25% trichloroacetic acid (TCA) overnight. Protein pellets were redissolved in 100 mM NH4HCO3 and the protein concentration was measured by Brad-ford assay. Histone proteins were derivatized by propionic anhydride and digested with trypsin for about 6 hours (Lin et al., 2012). Peptides were also derivatized by propionic anhydride and desalted by C18 Stage-tips. Histone peptides were loaded to a 75 µm I.D. × 15 cm fused silica capillary column packed with Reprosil-Pur C18-AQ resin (3 µm; Dr. Maisch GmbH, Germany) using an EASY- nLC 1000 HPLC system (Thermo Scientific, Odense, Denmark). The HPLC gradient was 2–35% solvent B (A = 0.1% formic acid in water; B = 0.1% formic acid in acetonitrile) in 40 min and from 35% to 98% solvent B in 20 minutes at a flow-rate of 300 nL/min. HPLC was coupled to an LTQ-Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany). Full MS spectrum (m/z 290−1400) was performed in the Orbitrap with a resolution of 60,000 (at 400 m/z), and the 10 most intense ions were selected for MS/MS performed with collision-induced dissociation (CID) with normalized collision energy of 35 in the ion trap. AGC targets of full MS and MS/MS scans are 1×106 and 1×104, respectively. Precursor ion charge state screening was enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 30 s. Lock mass calibration in full MS scan is implemented using polysiloxane ion 371.10123. Histone peptide abundances were calculated from the acquired raw data by EpiProfile program (in revision).
Immunoblot analysis
PBS Lysis Buffer (1% Triton X-100, 1mM DTT, in PBS) followed by 0.2 N HCl solution was used to prepare lysates for histone fraction of lymphoma/B220+ cells. RIPPA buffer was used to prepare whole cell lysates of OCI-LY7 cells. Immunoblot analyses were performed according to standard procedures. Membranes were probed with the indicated primary antibodies: H3K4me1 (Abcam, #ab8895), H3K4me2 (Millipore #07-030), H3K4me3 (Millipore #07-473), total H3 (abcam #ab1791), P-Tyr705-STAT3 (Cell Signaling #9145), total-STAT3 (Cell Signaling #12640), and SOCS3 (Cell Signaling #2932). Enhanced chemiluminescence was used for detection (ECL, Amersham).
Validation of KMT2D targets by quantitative real time PCR analyses
Total RNA from cells was extracted using TRIZOL (Invitrogen). Reverse transcription was performed using random primers and SuperScript III First Strand (Invitrogen #18080-400). Quantitative real time-PCR was performed using TaqMan Universal Master Mix (Applied Biosystems) in an 7900 HT Fast Real Time thermocycler (Applied Biosystem). The housekeeping gene used for input normalization of all the qRT-PCR data is β-actin. Taqman Gene Expression Assays used: Kmt2d (Mm02600438_m1), β-actin (#4352663), Socs3 (Mm00545913), Dusp1 (Mm00457274), Tnfaip3 (Mm00437121), Arid1a (Mm00473838), Fos (Mm00487425), Ikbkb (Mm01222247), Tnfrsf14 (Mm00619239), KMT2D (Hs00231606), SOCS3 (Hs02330328), TNFRSF14 (Hs00998604), TNFAIP3 (Hs00234713), ARID1A (Hs00195664), DUSP1 (Hs00610256), TRAF3 (Hs00936781), NR4A1 (Hs00374226), IKBKB (Hs00233287), DNMT3A (Hs01027166), ASXL1 (Hs00392415), ARID3B (Hs00356736), MAP3K8 (Hs00178297) and ACTB (#4352667).
Statistical methods
Sample sizes for comparisons between cell types or between mouse genotypes followed Mead's recommendations48. Samples were allocated to their experimental groups according to their pre-determined type (i.e mouse genotype) and therefore there was no randomization. Investigators were not blinded to the experimental groups. In the case Fig 1b only mice that developed lymphomas were considered; mice that didn’t developed lymphomas were censored and indicated with ticks in the Kaplan–Meier curves. Quantitative PCR data were obtained from independent biological replicates (n values indicated in the corresponding figure legends) Normal distribution and equal variance was confirmed in the large majority of data and, therefore, we assumed normality and equal variance for all samples. Based on this, we used the Student's t-test (two-tailed, unpaired) to estimate statistical significance. Survival in mouse experiments was represented with Kaplan-Meier curves, and significance was estimated with the log-rank test. For contingency analysis (Proportion of H3K4me1/me2 peaks) we used the Chi-square exact test.
Supplementary Material
Acknowledgements
We thank Elisa Oricchio (MSKCC), Viraj Shangvi (MSKCC), Michael Boice (MSKCC), Wendy Beguelin and María del Pilar Dominguez Rodriguez (Weill Cornell Medical College) for advice and reagents. Thanks to Itamar Niesvizky for assistance with CD40 stimulation experiments. Thanks to Elisa de Stanchina and all the members of MSK Antitumor assessment core for technical assistance with mice; Agnes Viale and Nicholas D. Socci for sequencing and data analysis; the MSK Laboratory of Comparative Pathology, MSK Flow Cytometry, and MSK Molecular Cytology cores. Herbert Hagenau for Southern blot analysis. A. O.-M. is supported by funding from The Leukemia & Lymphoma Society. H.G.W is supported by the American Cancer Society grant RSG-13-048-01-LIB, the Lymphoma Research Foundation, Cycle for Survival, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center, NIH grants RO1CA183876-01 and 1R01CA19038-01 and Core Grant (P30 CA008748). H.G.W is a Scholar of the Leukemia and Lymphoma Society. A.M.M. is supported by NIH R01CA187109, the Chemotherapy Foundation, and is a Scholar of the Bouroughs Wellcome Foundation. I.W.B. is supported by a Sass Foundation Post-doctoral fellowship award. B.A.G acknowledges funding from an NIH Innovator grant (DP2OD007447) from the Office of the Director, and NIH grant R01GM110174. A.S.H. is supported by NIH grant R01CA150265. The work by K.G Lab was supported by the Intramural Research Program of the NIDDK, NIH. The work by A.N. Lab was supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research, by a Department of Defense grant (BCRP DOD Idea Expansion Award, grant 11557134) and the Alex Lemonade Stand Foundation Award.
Footnotes
Accession codes:
The GEO numbers for the manuscript are the following:
GES67291: Mouse B220+ lymphoma H3K4me1/me2 ChIPseq and RNAseq
GES67314: KMT2D ChIPseq in OCI-LY7 lymhoma cells
GES67381: H3K4me1/me2 ChIPseq in OCI-LY7 and OCI-LY1 lymphoma cells
SRP056293: FL samples RNA-seq
SRP056292: targeted resequencing in FL samples
SRP056291: exome sequencing in FL samples
Author contributions
A.O.-M. designed and performed functional studies, analyzed data and wrote the manuscript; I.W.B. designed experiments, performed epigenetic studies, and analyzed data; A.C performed studies on Kmt2d−/− mice with the help of H.-T.C; H.P and O.E performed RNA-seq and ChIP-Seq analysis; Y.J. and O.E performed and analyzed exome sequencing and targeted re-sequencing in FL samples; C.Z. and M.J. provided technical assistance; D. H. and A.S.H. performed and analyzed KMT2D ChIP-seq in DLBCL cell lines; X. A. performed KMT2D expression analysis in B cell populations; I.M. helps with CD40/IgM experiments in cell lines; K. G. and J-E. L. generated the Kmt2df/f mice; D.E., D.W.S., C.H., A.M. and R.D.G. performed KMT2D sequencing and cell-of-origin determination in DLBCL samples; S.L., X.-J. and B.A.G. performed quantitative mass spectrometry analysis of lymphoma cell lines; R.S. performed pathological evaluation of mouse models; W. T. provided critical clinical samples; A. N. supervised the experiments on Kmt2d−/− mice. A.M.M. and H.G.W. designed and directed the study.
Author information The authors declare no competing financial interests.
Supplementary information includes seven supplementary figures and six supplementary tables.
References
- 1.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nature reviews. Immunology. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. The Journal of clinical investigation. 2012;122:3424–3431. doi: 10.1172/JCI63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu D, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herz HM, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JE, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell. 2014;53:859–866. doi: 10.1016/j.molcel.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egle A. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103:2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- 10.Oricchio E, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–564. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- 12.Santos MA, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman JD, et al. Immune abnormalities are a frequent manifestation of Kabuki syndrome. American journal of medical genetics. Part A. 2005;135:278–281. doi: 10.1002/ajmg.a.30722. [DOI] [PubMed] [Google Scholar]
- 14.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molavi O, et al. Gene methylation and silencing of SOCS3 in mantle cell lymphoma. Br J Haematol. 2013;161:348–356. doi: 10.1111/bjh.12262. [DOI] [PubMed] [Google Scholar]
- 16.Cheung KJ, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70:9166–9174. doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- 17.Guo C, et al. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4:2144–2153. doi: 10.18632/oncotarget.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarma V, et al. Activation of the B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apoptosis. The Journal of biological chemistry. 1995;270:12343–12346. doi: 10.1074/jbc.270.21.12343. [DOI] [PubMed] [Google Scholar]
- 19.Hsing Y, Hostager BS, Bishop GA. Characterization of CD40 signaling determinants regulating nuclear factor-kappa B activation in B lymphocytes. J Immunol. 1997;159:4898–4906. [PubMed] [Google Scholar]
- 20.Hollmann CA, Owens T, Nalbantoglu J, Hudson TJ, Sladek R. Constitutive activation of extracellular signal-regulated kinase predisposes diffuse large B-cell lymphoma cell lines to CD40-mediated cell death. Cancer Res. 2006;66:3550–3557. doi: 10.1158/0008-5472.CAN-05-2498. [DOI] [PubMed] [Google Scholar]
- 21.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjornsson HT, et al. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci Transl Med. 2014;6:256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nature reviews. Drug discovery. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Soong TD, Wang L, Melnick AM, Elemento O. Genome-wide detection of genes targeted by non-Ig somatic hypermutation in lymphoma. PLoS One. 2012;7:e40332. doi: 10.1371/journal.pone.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wacker SA, Houghtaling BR, Elemento O, Kapoor TM. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nature chemical biology. 2012;8:235–237. doi: 10.1038/nchembio.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajadhyaksha AM, et al. Mutations in FLVCR1 cause posterior column ataxia and retinitis pigmentosa. American journal of human genetics. 2010;87:643–654. doi: 10.1016/j.ajhg.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott DW, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 30.Wendel HG, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 31.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 32.Beguelin W, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mavrakis KJ, et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes & Development. 2008;22:2178–2188. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehlich A, Martin V, Muller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Current biology : CB. 1994;4:573–583. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 36.Gostissa M, et al. Conditional inactivation of p53 in mature B cells promotes generation of nongerminal center-derived B-cell lymphomas. Proc Natl Acad Sci U S A. 2013;110:2934–2939. doi: 10.1073/pnas.1222570110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic acids research. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ci W, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature protocols. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cover TM, Thomas JA. Elements of information theory. Hoboken, N.J: Wiley-Interscience; 2006. [Google Scholar]
- 43.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaffer AL, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunological reviews. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 45.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 46.Mavrakis KJ, Wendel HG. Translational control and cancer therapy. Cell Cycle. 2008;7:2791–2794. doi: 10.4161/cc.7.18.6683. [DOI] [PubMed] [Google Scholar]
- 47.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods in enzymology. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Festing MFW Laboratory Animals Ltd. The design of animal experiments : reducing the use of animals in research through better experimental design. London: Royal Society of Medicine; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.