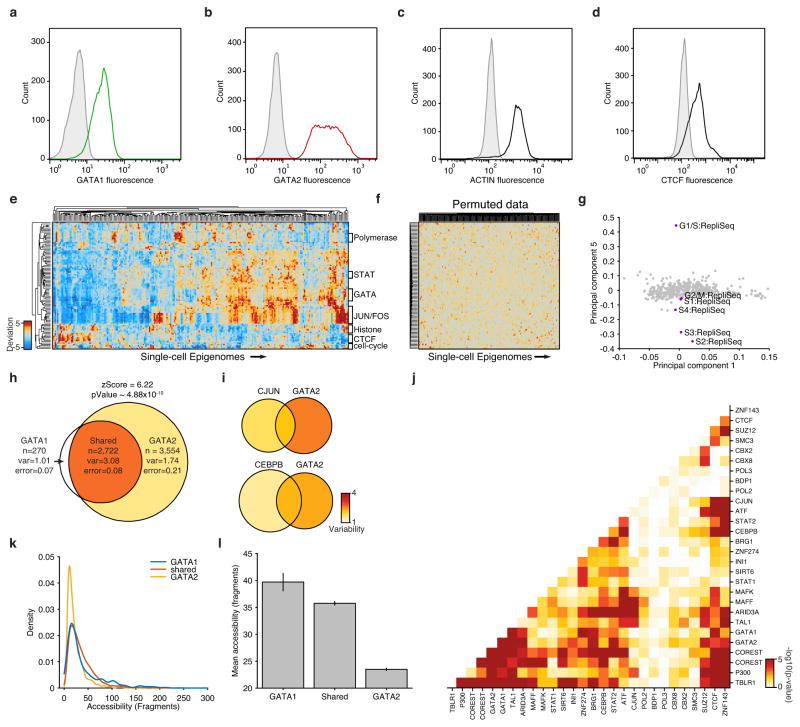
Extended Data Figure 6. Characterization of high-variance trans-factors in K562 cells.
(a–d) Distribution of (a) GATA1, (b) GATA2, (c) actin and (d) CTCF fluorescence observed by flow cytometry. Distributions in grey depict isotype controls. (e) Bi-clustered heat map of single cell deviations as observed within K562 cells (N=239). Labels on right identify co-clustering of related factors. (f) Bi-clustered heat map of single-cell deviations observed from permuted data. (g) Projection of factor loadings onto principal component 1 versus 5 from principal component (PC) analysis of heatmap from Fig. 2d. Factor loadings do not vary along PC5, while peaks associated with regions with different replication timings (RepliSeq) have strong variation along this axis. Venn-diagrams showing variability of (h) GATA1 and/or GATA2, (i) CJUN and/or GATA2 and CEBPB and/or GATA2 (co-) occurring ChIP-seq sites. (j) -log10(p-values) of calculated changes in co-occurring ChIP-seq sites shown in Figure 2e. (k) Distribution of accessibility among GATA1 only, GATA2 only, and shared sites. (l) Mean accessibility from GATA1 only, GATA2 only, and shared sites in (k), error bars represent 1 standard deviation generated by bootstrapping ChIP-seq peaks.