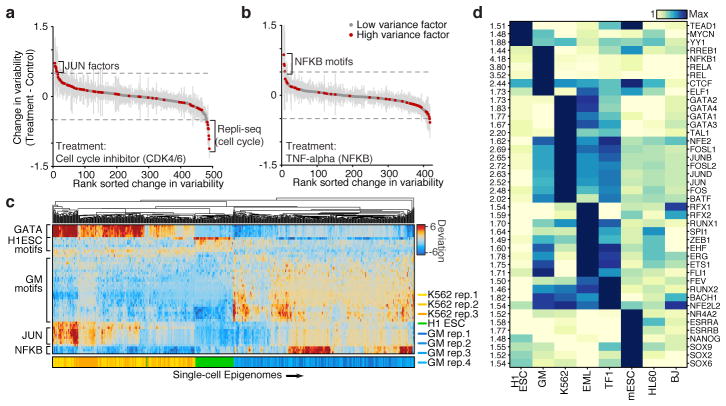
Figure 3. Cell type specific epigenomic variability.
Change of cellular variability due to chemical perturbations using (a) CDK4/6 cell-cycle inhibitor (K562) or (b) TNF-alpha stimulation (GM12878), error bars (shown in grey) represent 1 standard deviation of bootstrapped cells across the two conditions. (c) Heat map of deviations from expected accessibility signal across trans-factors (rows) and of single cells (columns) from 3 cell types. Bottom color map represents assignment classification from hierarchical clustering. (d) Variability associated with trans-factor motifs across 7 cell types. Each row is normalized to the maximum variability for that motif across cell types (shown left).