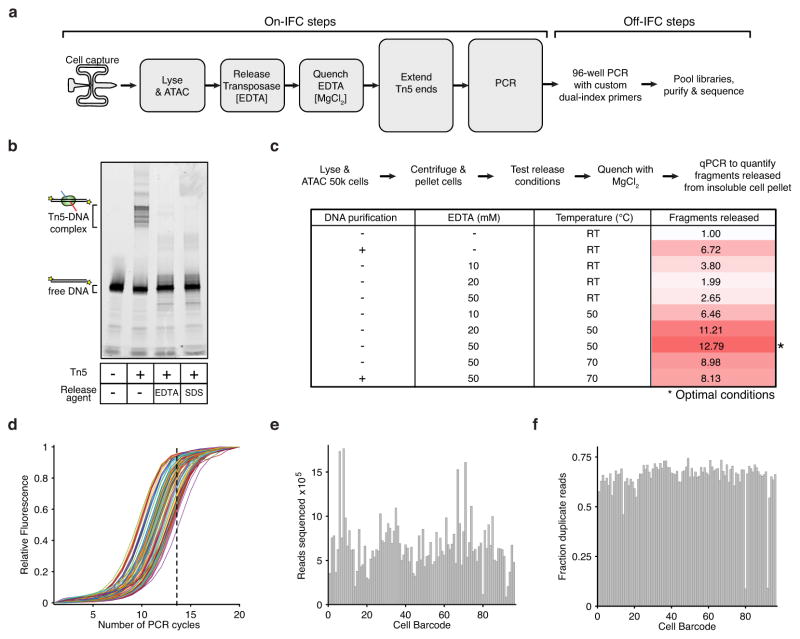
Extended Data Figure 1. Methods development for assaying single epigenomes.
(a) scATAC-seq workflow for steps performed both on and off Fluidigm’s integrated fluidics circuit (IFC). (b–c) The development of an efficient Tn5 release protocol designed to permit downstream enzymatic reactions without DNA purification. (b) An in vitro electrophoretic mobility gel shift assay using a fluorescently labeled PCR product (lane 1), showing a stable Tn5-DNA complex (lane 2) dissociated with 50 mM EDTA (lane 3) or 0.1% SDS (lane 4). (c) Workflow and associated table of conditions used to optimize release protocol, showing conditions that markedly improve fragment yield over no release conditions or purifying DNA (Qiagen MinElute). Fragments released represents the fold gain in library diversity, as measured by quantitative PCR (qPCR). (d) qPCR fluorescence traces of 96 libraries generated using scATAC-seq. For all subsequent libraries we used a total of 14 PCR cycles (dotted line). (e,f) A bar plot of per-cell library (e) sequencing depth and (f) fraction of duplicate reads, showing each library was sequenced to varying depths to a similar fraction of duplicate reads.