Abstract
Developing technologies for efficient and scalable disruption of gene expression will provide powerful tools for studying gene function, developmental pathways, and disease mechanisms. Here we develop CRISPR interference (CRISPRi) to repress gene expression in human induced pluripotent stem cells (iPSCs). CRISPRi, in which a doxycycline-inducible deactivated Cas9 is fused to a KRAB repression domain, can specifically and reversibly inhibit gene expression in iPSCs and iPSC-derived cardiac progenitors, cardiomyocytes, and T lymphocytes. This gene repression system is tunable and has the potential to silence single alleles. Compared with CRISPR nuclease (CRISPRn), CRISPRi gene repression is more efficient and homogenous across cell populations. The CRISPRi system in iPSCs provides a powerful platform to perform genome-scale screens in a wide range of iPSC-derived cell types, and to dissect developmental pathways and model disease.
Introduction
To understand the biological roles of genes in development and disease, we must decipher the relationships between genotype and phenotype. Until recently, RNAi has been the most commonly used loss-of-function tool to study human biology (Boettcher and McManus, 2015). However, RNAi suffers from off-target effects and incomplete silencing of the desired gene (Jackson et al., 2003; Kim et al., 2013b; Krueger et al., 2007). Alternatively, programmable nucleases, such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), allow more precise gene editing in model organisms, particularly in mammalian and human systems (Gaj et al., 2013; Kim and Kim, 2014). While ZFNs and TALENs are efficient tools for targeting single alleles, they cannot be easily used for library-scale loss-of-function studies.
In 2012, clustered regularly interspaced short palindromic repeat (CRISPR) technology emerged as a new tool for gene editing. This technology is a microbial adaptive-immune system that uses RNA-guided nucleases to recognize and cleave foreign genetic elements (Doudna and Charpentier, 2014; Wiedenheft et al., 2012). The recently engineered CRISPR/Cas9 system consists of two components: a single-chimeric guide RNA (gRNA) that provides target specificity and a CRISPR-associated protein (Cas9) that acts as a helicase and a nuclease to unwind and cut the target DNA (Cong et al., 2013; Mali et al., 2013). In this system, the only restriction for targeting a specific locus is the protospacer adjacent motif (PAM) sequence (“NGG” in the case of SpCas9) (Doudna and Charpentier, 2014).
CRISPR nuclease (CRISPRn) has been used for genome-scale screens to identify essential genes for cell viability in cancer and embryonic stem cells (Shalem et al., 2013), and human leukemic cell lines (Wang et al., 2013, 2015). However, CRISPRn may not be the most robust system for loss-of-function studies because it is limited by the number of cells within a population that do not produce knockout phenotypes (González et al., 2014). In addition, partial loss- or gain-of-function phenotypes can be generated by Cas9-induced in-frame insertion/deletions (INDELs) and hypomorphic alleles (Shi et al., 2015), which can obscure the readout.
The nuclease deactivated version of Cas9 (dCas9) blocks transcription in prokaryotic and eukaryotic cells (known as CRISPR interference; CRISPRi) (Qi et al., 2013). More recently, dCas9 was fused to the Krüppel-associated box (KRAB) repression domain (dCas9-KRAB), producing a more efficient transcriptional interference (Gilbert et al., 2013, 2014; Kearns et al., 2014). To further this effort, we aimed to use CRISPRi technology to efficiently repress genes to study early differentiation, and to model disease with human induced pluripotent stem cells (iPSCs) (Takahashi et al., 2007).
iPSCs are well suited to study early embryonic development and disease since they can produce different functional cell types in vitro (Sterneckert et al., 2014). Early embryonic development consists of a series of accurately timed events that affect gene activation and repression (Bolouri and Davidson, 2003). Therefore, precisely regulating the timing and dosage of transcription factors critically affects embryonic development (McFadden et al., 2005; Takeuchi et al., 2011), and dysregulation in the timing and dosage of transcripts can lead to disease development (Theodoris et al., 2015). In this study, we compared inducible CRISPR systems for gene knockout (using Cas9) or knockdown (using dCas9-KRAB) to enable temporal control over loss-of-function phenotypes in iPSCs and differentiated cell types.
Results
Generation of CRISPRi and CRISPRn iPSC Lines
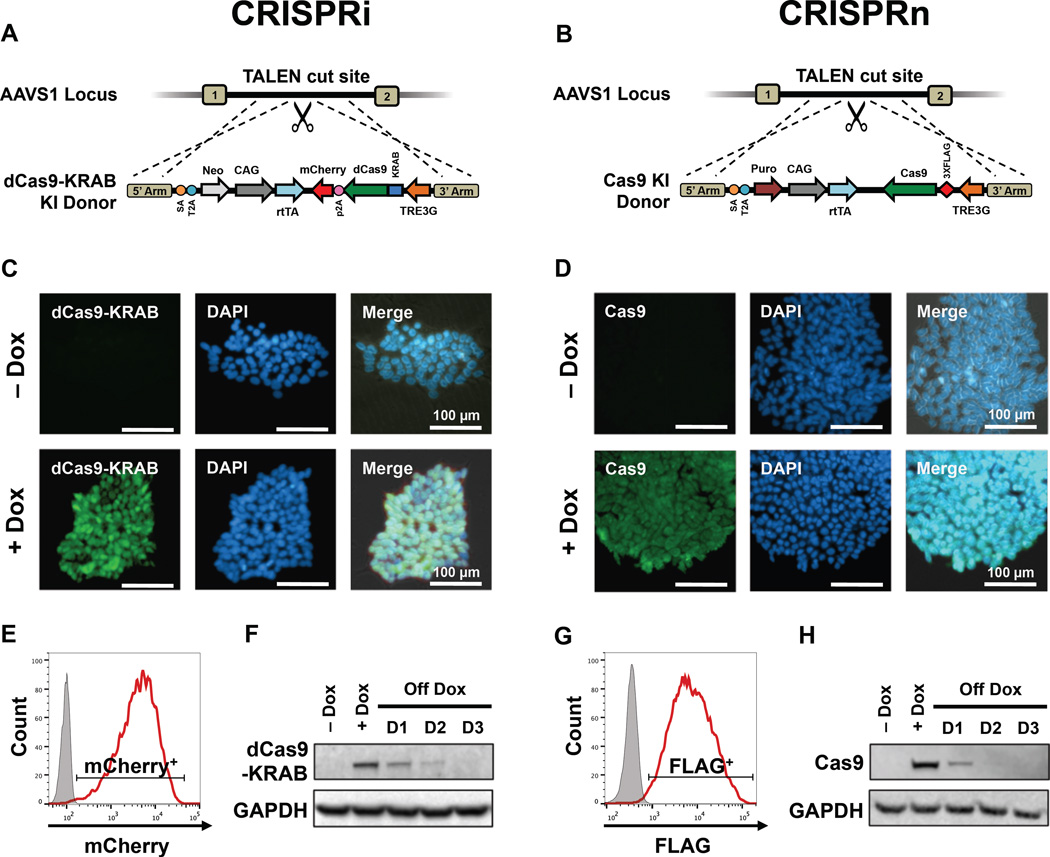
For loss-of-function studies, we generated multiple independently derived stable CRISPRi and CRISPRn human iPSC clones in two genetic backgrounds, wild-type B (WTB) and wild-type C (WTC) (Miyaoka et al., 2014). In separate targeting events, the CRISPRi and CRISPRn constructs (Table S1) were integrated into the AAVS1 locus of WTB and WTC iPSCs using TALEN-assisted gene-trap approach (Figures 1A, 1B and S1). Transgenes integrated at the AAVS1 locus remain transcriptionally active in both iPSCs and differentiated cell types (Hockemeyer et al., 2011; Lombardo et al., 2011). We have generated several different versions of the CRISPRi system, that are either inducible or constitutive: the inducible CRISPRi (Gen1 and Gen2) clones express dCas9-KRAB (KRAB domain fused at the N-terminus) from the inducible TetO promoter, while the constitutive CRISPRi clones (Gen3) express dCas9-KRAB under the constitutively-active CAG promoter. The CRISPRn (Gen1) clones express Cas9 under the inducible TetO promoter (Figure S1).
Figure 1. Generation and characterization of inducible CRISPRi and CRISPRn iPSCs.
(A, B) Schematic overview of the strategy for TALEN-mediated targeting to the AAVS1 locus to generate the CRISPRi and CRISPRn iPSC lines. The doxycycline-controlled transcriptional activator (rtTA) is driven by a strong constitutive promoter (CAG). The third-generation doxycycline-response element (TRE3G) drives transcription of either Cas9 (CRISPRn) or dCas9-KRAB-P2A-mCherry (CRISPRi) and is oriented in the opposite direction of the transactivator to ensure no leaky expression without doxycycline treatment. (C, D) Immunostaining of CRISPRi and CRISPRn colonies before and after 48 h of doxycycline treatment with an antibody against Cas9 (green). Nuclei are stained with DAPI (blue). All nuclei showed expression of dCas9-KRAB or Cas9 after adding doxycycline. (E, G) Flow cytometry analysis of CRISPRi and CRISPRn iPSC lines before and after 48 h of doxycycline treatment. Doxycycline treatment of CRISPRi and CRISPRn produced expression of mCherry and FLAG in all cells, respectively. The doxycycline-untreated sample is plotted in grey. (F, H) CRISPRi and CRISPRn iPSC lines were treated with doxycycline (2 µM) for 24 h, which was then removed to measure the protein half-life of dCas9-KRAB and Cas9. Total protein was extracted from samples and analyzed by western blot with antibodies against Cas9 and GAPDH as a loading control. Both the CRISPRi and CRISPRn clones express dCas9-KRAB and Cas9 at similar levels after doxycycline treatment and the half-life of both proteins was approximately 12 h in iPSCs. Scale bars = 100 µm.
The average efficiency of forming stable clones was ~350 colonies per million iPSCs transfected with AAVS1 TALENs and donor plasmid (data not shown). From each condition, multiple independent colonies were isolated and expanded. A subset of the stable colonies from each targeting vector was screened using junction PCR. Two putative colonies from each targeting event were further characterized by stably introducing an OCT4-specific gRNA and performing knockdown or knockout assays with immunofluorescence and western blot analysis. All putative CRISPRi clones containing an OCT4-specific gRNA showed efficient knockdown (>95%) of OCT4 in bulk populations, while a significant fraction of the CRISPRn cells remained OCT4-positive (~30–40%) in bulk populations containing OCT4-specific gRNA (Figure S1). One clone each from CRISPRi and CRISPRn (Gen1 lines in the WTC genetic background) were subsequently used as lead clones for further studies.
To enable non-invasive and high-throughput phenotypic analysis in iPSC-derived cardiomyocytes (iPS-CM), we performed a second targeting event that introduced the green fluorescent calcium-modulated protein 6 fast type (GCaMP) calcium sensor (Chen et al., 2013) into the other AAVS1 locus of the CRISPRi cell line. The GCaMP transgene is driven off the strong, constitutive CAG promoter (Figure S1). We found that CRISPRi iPSCs could differentiate into iPS-CM, so that we could measure calcium transients based on the GCaMP-fluorescent intensity (Video S1) (Huebsch et al., 2015). Lead CRISPRi and CRISPRn iPSCs were karyotypically normal (Figures S2A and S2B) and expressed pluripotency markers, as expected (Figures S2C and S2D).
RNA-Seq analysis indicated that expression of dCas9-KRAB or Cas9 was undetectable in the absence of doxycycline, and addition of doxycycline without any gRNA resulted in robust selective induction of dCas9-KRAB or Cas9, while the rest of the transcriptome remained virtually unchanged (Figures S2E and S2F). Furthermore, the RNA-Seq data suggests that the addition of the KRAB domain has no detectable off-target effects as compared to expression of Cas9 alone. Remarkably the one gene that appeared to be upregulated upon doxycycline induction (without gRNA) was the same gene (Vimentin; VIM) for both CRISPRi and CRISPRn cells (Figures S2E and S2F). Since the same gene is up-regulated for CRISPRi and CRISPRn cells, we suspect it may represent an off-target activity of the doxycycline-induced transactivator. Importantly, our experiments suggest that the expression of dCas9-KRAB alone has no additional changes on gene expression.
We also expressed dCas9-KRAB and Cas9 by continuously culturing CRISPRi and CRISPRn lines with doxycycline for 3 weeks (four passages). With this long-term treatment, we observed no cytotoxicity, decrease in proliferation, or change in morphology in these cells (Figures S2G and S2H). Using droplet digital PCR (ddPCR)-based copy number assay, we measured the number of integration events (Figure S2I). We further validated on-target integration sites on the lead CRISPRi and CRISPRn clones with junction PCR (Figure S2J) and verified their sequences (data not shown).
To further ensure there was no leaky expression of the single doxycycline-inducible vector, we measured the protein levels of dCas9-KRAB and Cas9 in iPSCs. With immunostaining, flow cytometry and western blots did not detect dCas9-KRAB or Cas9 protein without doxycycline in either CRISPRi or CRISPRn iPSCs, indicating that the TetO promoter has high fidelity in the AAVS1 locus. After doxycycline treatment, all cells in the CRISPRi and CRISPRn lines expressed dCas9-KRAB or Cas9 within 48 h, respectively (Figures 1C–H). dCas9-KRAB and Cas9 were expressed at similar levels after induction and both proteins rapidly degraded after removing doxycycline (Figures 1F, 1H, S2K). These data showed that dCas9-KRAB and Cas9 expression could be tightly regulated with the TetO promoter, which would support studies that rely on precisely timing gene knockdown or knockout.
Comparison of Loss-of-Function between CRISPRi and CRISPRn
To compare CRISPRi and CRISPRn for loss-of-function studies, we designed a gRNA that targets the first exon of NANOG, a transcription factor necessary for maintaining the pluripotency network. We selected NANOG as our first target gene because its deficiency is sufficient to give an immediate readout, as indicated by a clear loss of pluripotent cell morphology (Hayashi et al., 2015). In general, Cas9 can disrupt gene function at any given exon (Doench et al., 2014) while dCas9-KRAB knocks down gene expression only when gRNAs are targeted to the transcription start site (TSS) (Gilbert et al., 2014). Hence, for this comparative study, we used the same gRNA sequence for both CRISPRi and CRISPRn. Here, we introduced a gRNA targeting 358 bp downstream of the NANOG TSS (142 bp into exon 1 of NANOG) into the CRISPRi and CRISPRn clones and selected subclones (as described in Materials and Methods). We then treated multiple independent subclones of CRISPRi and CRISPRn iPSCs containing the NANOG gRNA-expression vector (as indicated by mKate2 expression) with doxycycline (Figure 2).
Figure 2. Comparison of the efficiency of CRISPRi knockdown and CRISPRn knockout.
Immunostaining of representative (A) CRISPRi and (B) CRISPRn stable clones, each containing the same gRNA targeting the first exon of NANOG (NANOG g+358). After 7 days of doxycycline treatment, NANOG expression (green) was completely lost in all CRISPRi clones, but showed a variegated pattern of knockout in multiple independent CRISPRn clones. The mKate2 signal indicates the presence of the gRNA-expression vector in all cells within the clone. Nuclei are counterstained with DAPI. Western blot and flow cytometry analyses of (C, E) CRISPRi and (D, G) CRISPRn stable clones that contain the same gRNA against the first exon of NANOG. With CRISPRi, NANOG expression was uniformly decreased during doxycycline treatment and did not increase thereafter; however, with CRISPRn, the percentage of NANOG-positive cells fluctuates over the period of doxycycline treatment. Even after 12 days of continuous doxycycline treatment, ~30% of the population stained positive for NANOG. Genomic DNA was extracted from (F) CRISPRi and (H) CRISPRn stable lines containing a gRNA against NANOG before and after continuous doxycycline treatment for up to 17 days and subjected to sequencing. Red, out-of-frame INDELs; orange, in-frame INDELs; green, non-mutated alleles. Even after 12–17 days of continuous doxycycline treatment, 50–70% of sequenced alleles from CRISPRn contained no mutation, and 30–50% of mutated alleles were in-frame INDELs. No mutations were observed in either CRISPRi or CRISPRn without doxycycline, and the CRISPRi clones did not contain any mutations after doxycycline treatment. The total number of sequenced colonies is listed below each pie graph. Scale bars = 500 µm.
With CRISPRi, we found that NANOG expression was completely lost (>99%) in multiple independent iPSC subclones after doxycycline treatment (Figures 2A, 2C, 2E, S3A and S3C). However, with CRISPRn, only 60–70% of the cells lost NANOG expression in multiple independent subclones post-doxycycline induction (Figure 2B, 2D, 2G, S3B and S3D). Next, we extracted genomic DNA from NANOG gRNA-containing CRISPRi and CRISPRn iPSCs and performed sequence analysis. As expected, we found that CRISPRi iPSCs did not harbor any mutations in the NANOG locus pre- or post-doxycycline treatment (Figure 2F). However, with CRISPRn, after 12–17 days of continuous doxycycline treatment, among the mutated alleles, 30–50% of the sequences contained in-frame INDELs at the cut site (a total of 77 sequenced clones) (Figure 2H).
To further compare CRISPRi with CRISPRn, we targeted another pluripotency transcription factor, OCT4, with two independent gRNAs. Similar to our findings with NANOG, OCT4 was completely knocked down in independent CRISPRi clones expressing the gRNA vector after doxycycline treatment (Figure S3E). In contrast, the attempted knockout of OCT4 with CRISPRn again yielded incomplete effects (Figure S3F). These findings were also replicated in a completely different iPSC line (WTB genetic background; CRISPRi Gen1B and CRISPRn Gen1B) (Figures S1D and S1F). We analyzed the genomic DNA of CRISPRn cells after 14 days of continuous doxycycline treatment, and found 30–40% of the mutated alleles had in-frame INDELs (a total of 91 sequenced clones) (Figure S3G). These results suggested that, in the context of targeting pluripotency factors, CRISPRi more rapidly generates loss-of-function phenotypes in bulk populations than CRISPRn. CRISPRi caused a complete loss of transcript expression and rapid cell differentiation when targeting NANOG and OCT4 within 5–7 days of knockdown initiation. With CRISPRn, even after approximately 2 weeks of doxycycline treatment, a significant fraction (30–40%) of the cells remained NANOG- and OCT4-positive and maintained their pluripotency. Therefore, we focused on using CRISPRi as a loss-of-function tool in subsequent experiments.
CRISPRi Is Most Effective near the TSS
To further test the efficacy of gRNAs in CRISPRi, we designed multiple gRNAs that target near the TSS of OCT4. With flow cytometry assays for OCT4 staining (Figure 3A), we found that most gRNAs targeting near the TSS (approximately − 150 bp to +150 bp around the TSS in this study) were highly effective at gene knockdown, but gRNAs targeting significantly (>700 bp) downstream of the TSS were not. This result agrees with previous data (Gilbert et al., 2014) and suggests that CRISPRi primarily blocks transcription at initiation, which reduces the likelihood of off-target effects from transcript interference elsewhere in the genome. Following these design criteria, for subsequent gene targets we designed gRNAs to target near the TSS.
Figure 3. CRISPRi knockdown is efficient in iPSCs.
(A) Efficiency of gRNA knockdown based on proximity to the transcription start site (TSS). The binding location of each gRNA is indicated relative to the TSS of the OCT4 locus and whether it targets the template (T) or non-template (NT) strand. Only gRNAs targeting near the TSS (approximately ± 150 bp) effectively knocked down OCT4. (B) TaqMan qPCR analysis of stable iPSCs containing gRNA against the gene of interest showed greater than 90% knockdown efficiency after 7 days of doxycycline induction in different endogenous genetic loci. (C) Immunostaining of stable clones containing a single gRNA against the gene of interest (OCT4, SOX2, NANOG, and BAG3). After 7 days of doxycycline treatment, there was a complete knockdown of the protein of interest (green). As expected, DAPI staining revealed that knocking down OCT4, NANOG and SOX2 resulted in loss of pluripotency and clear morphological changes. Also, knocking down BAG3 did not cause a loss of pluripotent morphology, as indicated by the distinct and round colony edges.
CRISPRi Efficiently Knocks Down a Broad Range of Genetic Loci
To test the efficiency of CRISPRi across a broad range of genetic loci in both iPSCs and differentiating/differentiated cell types, we designed gRNAs against a total of nine genomic loci. The loci included core pluripotency transcription factors (OCT4, NANOG, and SOX2), kinases (ROCK1 and GSK3-β), a cardiac mesoderm-transcription factor (MESP1), and cardiac disease-associated genes (BAG3, MYBPC3, and HERG). Except for MESP1 (expressed only transiently in cardiac mesoderm cells) and MYBPC3 (expressed only in cardiomyocytes), all other genes are expressed in iPSCs at different levels. We generated populations of CRISPRi iPSCs containing stably integrated gRNA-expression constructs. We then cultured these stable polyclones or clonal populations either with or without doxycycline for at least 7 days.
Three to five gRNAs were designed to target near the TSS of each gene, and initially were tested individually in polyclonal populations. Approximately half of the tested gRNAs were active in polyclonal populations with a silencing activity of over 70% (Figure S4A). We did not observe a difference in the knockdown efficiency between gRNAs targeting either the template or non-template strands (Figures 3A, S4A and S4B). The most active gRNA-containing polyclonal line was further passaged and subcloned for more detailed knockdown analysis. Using the most active gRNA, we achieved 90–99% knockdown of the gene of interest in a selected population of iPSCs after doxycycline treatment (Figure 3B). As expected, when we subcloned polyclonal populations via single-cell cloning, we observed a higher percentage of transcriptional knockdown. With immunofluorescence analysis we found that iPSC clones expressing gRNAs against OCT4, NANOG, SOX2, and BAG3 showed complete loss of target protein expression 7 days after doxycycline induction. In cells expressing gRNAs against the core pluripotency transcription factors OCT4, NANOG, and SOX2, we observed clear morphological changes and a loss of pluripotency after doxycycline induction; however, loss of a non-pluripotency gene (BAG3) did not affect pluripotent morphology (Figure 3C).
Using Gen1 CRISPRi knockin vector, we targeted non-iPSCs with s different genetic background to determine how broadly this technology can be applied to other cell types. A T-lymphocyte (CEM) CRISPRi line was generated, as described in Materials and Methods. Similar to the iPSC experiments, gRNAs was introduced to the stable CEM CRISPRi cell line and were cultured either in the presence or absence of doxycycline for 10 days. Three gRNAs were tested to knockdown CD4 in CEM-CRISPRi cells and all showed greater than 70% knockdown efficiency in polyclonal populations (Figure S4B). The most active gRNA-containing polyclone was subcloned and three independent clonal lines were isolated and assayed for knockdown, where greater than 95% knockdown efficiency was observed (Figure S4C). These results clearly demonstrate the doxycycline-inducible CRISPRi vector system is highly versatile and transportable to other cell lines, and shows high efficiency of knockdown across a range of cell types and genetic loci.
CRISPRi Knockdown Is Reversible, Tunable, and Can Be Allele Specific
GCaMP is a calcium-sensitive modified GFP and thus, can be used as a fluorescent reporter under steady-state level of cytoplasmic Ca2+ (Apáti et al., 2013). Using GCaMP (driven off the strong constitutive promoter, CAG), we monitored the green fluorescence signal in iPSCs to determine if we could knock down GCaMP and then reverse its expression by removing doxycycline from the culture. We found that adding doxycycline for 7 days knocked down GCaMP expression by 98%, which was completely restored after removing doxycycline for 14 days (Figure 4A). Similarly, we targeted the BAG3 endogenous locus, we achieved efficient transcript knockdown post-doxycycline treatment and BAG3 expression was fully restored after doxycycline withdrawal (Figure 4B). These findings indicate that CRISPRi knockdown is fully reversible in iPSCs.
Figure 4. CRISPRi knockdown is reversible and tunable.
A CRISPRi clone containing gRNA against the GCaMP transgene (GCaMP g+56) and endogenous BAG3 locus were used to test the knockdown efficiency and reversibility of the CRISPRi system in iPSCs. (A) Flow cytometry analysis of GCaMP expression showed that after 7 days of doxycycline induction, GCaMP was knocked down by ~99%, and was completely restored after doxycycline withdrawal for 14 days. (B) Using TaqMan qPCR, BAG3 transcript levels were knocked down to nearly undetectable levels, and expression was restored after doxycycline withdrawal. (C) Schematic diagram of the GCaMP-expression vector in which the GCaMP open reading frame (ORF) is driven off the CAG promoter. The locations of three gRNAs (g+24, g+56, and g+91) are schematically highlighted on the GCaMP ORF. The coordinates of GCaMP gRNA are based on the translation start site. pA, poly A signal. (D) Three stable CRISPRi colonies, each containing a different gRNA against GCaMP, were selected using blasticidin and cultured with doxycycline for 10 days. The percentage of GCaMP-positive cells for each gRNA-containing clone was plotted as a function of time based on flow cytometry analysis. Variable levels of GCaMP knockdown (~30%, ~50%, and ~99%) were achieved with different gRNA sequences. Error bars = standard deviation. N=1–3 technical replicates for each time point. (E) Flow cytometry plots of GCaMP fluorescence of stable CRISPRi clones on day 10 of doxycycline treatment. Using different gRNAs that target near the same region, variable levels of knockdown can be achieved. A scramble gRNA-containing CRISPRi and a GCaMP-negative iPSC population are displayed as controls. (F) Partial schematic diagram of the OCT4 locus marked with the location of the TSS and two gRNA-binding locations. Asterisk, a single-nucleotide polymorphism (SNP); green box, exon 1; grey box, 5’ untranslated region (5’ UTR). (G) Three stable CRISPRi colonies, two with different gRNAs against OCT4 and one with a scrambled control, were selected with blasticidin. Stable clones that contain either a scramble gRNA, a gRNA that targets a PAM sequence containing a SNP (OCT4 g−4), or a gRNA that does not target a SNP (OCT4 g+22) were treated with doxycycline. The percentage of the maximal median intensity of OCT4 staining for each gRNA-containing clone is plotted as a function of time by flow cytometry analysis. Complete loss of OCT4 expression (>98% knockdown) was observed after 7 days of doxycycline induction only when both alleles were targeted using OCT4 g+22. While using OCT4 g−4, which targets only one OCT4 allele (due to SNP in the PAM sequence), a gradual loss of OCT4 staining intensity is observed over time (down by ~40% by day 7). Error bars = standard deviation. N=1–3 technical replicates for each time point. (H) Flow cytometry plots of OCT4 staining on day 7 of doxycycline treatment. Dashed lines highlight the loss of OCT4-staining intensity (~40%) when using OCT4 g−4 compared to the scramble control. By targeting only one allele of OCT4, the OCT4-staining intensity homogeneously shifts (while remaining OCT4-positive), indicating that each cell experiences approximately the same level of knockdown. Note that the x-axis is a log-scale of OCT4 intensity. Differentiated iPSC-derived fibroblasts (OCT4− Cntrl) and a non-doxycycline-treated (–Dox) sample are displayed as controls.
To determine if we could achieve variable levels of knockdown with different gRNA sequences, we tested two additional gRNAs targeting GCaMP (g+24 and g+91) (Figure 4C). These gRNAs knocked down GCaMP expression by only ~30% and ~50% as measured by flow cytometry (Figures 4D and 4E). Therefore, by changing the location of the gRNA binding site, we can tune the level of knockdown when trying to mimic haploinsufficiency or reduced protein levels (rather than complete loss-of-function). In addition, we tested whether the knockdown level is tunable by titrating the doxycycline concentration. Careful titration of the doxycycline concentration enabled homogenous modulation of GCaMP expression (Figure S5).
We next sought to further test the tunability of knockdown with CRISPRi. We determined if we could use single-nucleotide polymorphisms (SNP) to specifically target one allele for knockdown to achieve a heterozygous-like state. In our CRISPRi iPSCs, there is a SNP near the TSS of OCT4. Thus, we designed a gRNA in which the heterozygous SNP is located in the PAM sequence (AGG versus AGA). Because an “NGG” sequence is required for dCas9 to target DNA, we could selectively target only one OCT4 allele (Figure 4F). After doxycycline induction, we found that the iPSC population carrying the SNP-specific OCT4 gRNA (OCT4 g−4) remained OCT4-positive (~97%) by flow cytometry analysis. However, the median intensity of OCT4 staining was reduced by ~40% after 7 days of doxycycline treatment, implying that OCT4 expression was homogeneously reduced in all cells and not just a fraction of them (Figures 4G and 4H). We confirmed this finding with immunocytochemistry and Taqman qPCR (data not shown).
CRISPRi Knockdown is Highly Specific
To assess the specificity of CRISPRi targeting, we designed a gRNA that targets the GCaMP transgene, since its silencing should have few downstream transcriptional and cellular consequences. Indeed, expression of the GCaMP transcript was over 30-fold lower in the doxycycline-treated condition, while few other endogenous transcripts changed expression level with the exception of VIM as previously discussed (Figure 5A).
Figure 5. RNA-Seq and TaqMan qPCR analysis.
(A) RNA-sequencing RPM (reads per million) are plotted for CRISPRi cells stably expressing a gRNA targeting the GCaMP transgene (GCaMP g+56) cultured in the absence or presence of doxycycline. CRISPRi knockdown is specific to the GCaMP transcript, and few off-target transcriptional changes are observed. Data represent two independent biological replicates. (B) Heat map of TaqMan qPCR of stable clones containing a single gRNA against the gene of interest (OCT4, NANOG, and SOX2) as a function of days after doxycycline treatment. Analysis shows that by day 3, over 80% of the target transcript is depleted. Three housekeeping genes (18S, GAPDH, and UBC) were used to measure relative transcript levels. Each data point is an average of 2–4 technical replicates. TaqMan probes are listed in Table S5.
CRISPRi to Promote iPSC Differentiation
To show that our system can release iPSCs from their pluripotent state to promote differentiation, we tested the efficiency of CRISPRi in knocking down core pluripotency transcription factors (OCT4, SOX2, and NANOG) without adding small molecules or cytokines to the mTeSR media. We targeted gRNA against these genes and performed a time-course analysis of a selected number of transcripts by TaqMan qPCR (Figure 5B). We found that knocking down these target transcripts caused cell differentiation, as indicated by morphological changes and transient expression of the lineage-specific transcript T (mesoderm marker), and expression of PAX6 (neuronal progenitor marker). After 3 days of doxycycline treatment, over 80% of the target transcript was depleted, indicating that CRISPRi can precisely and temporally control efficient knockdown of the transcript of interest.
CRISPRi Knockdown in Cardiac Mesoderm and iPS-CMs
To determine if loss-of-function approaches using CRISPRi can be applied in differentiated cell types, we targeted the cardiac mesoderm-specific transcription factor (MESP1) and two known cardiac-related disease-causing genes (MYBPC3 and HERG). We established stable iPSC polyclonal lines containing gRNA against these three genes and differentiated them into cardiac mesoderm or iPS-CM as described in Materials and Methods (Figures S6A and S6B). Using a gRNA against these genes, MESP1 was knocked down by ~90% in cardiac progenitor cells, and MYBPC3 and HERG by ~90% and 60% respectively, in lactate-purified iPS-CM (Figure 6A). With western blots and immunocytochemistry, we observed ~90% MYBPC3 protein knockdown on day-35 lactate-purified iPS-CM (Figures 6B and 6C).
Figure 6. CRISPRi knockdown in differentiated cell types and cardiac-disease modeling.
(A) Using CRISPRi, MESP1 was knocked down by ~90% in polyclonal cardiac progenitors, and MYBPC3 and HERG were knocked down by ~90% and 60% in polyclonal iPS-CMs, respectively. (B) Immunostaining of day-35 lactate-purified iPS-CMs stained with antibodies against MYBPC3 (green) and ACTN2 (red). Using CRISPRi knockdown, loss of MYBPC3 was observed in over 85% of analyzed cells in a polyclonal population. Nuclei were counterstained with DAPI. Scale bar = 100 µm. (C) Western blot of day-35 lactate-purified iPS-CMs with antibodies against MYBPC3, ACTN2, and GAPDH. Using CRISPRi, MYBPC3 protein was knocked down by ~90%. (D) GCaMP fluorescence in iPS-CMs containing gRNA against HERG and cultured in doxycycline (red). Recordings show a prolonged beat duration compared to untreated controls (green). (E) Quantified ratio of the downstroke-to-upstroke duration of doxycycline-treated iPS-CMs shows a significant difference in untreated iPS-CMs containing a gRNA against HERG, but not in iPS-CMs containing gRNA against OCT4 (negative control). (F) Patch-clamp recordings from single iPS-CMs show prolonged action potential durations in doxycycline-treated samples containing HERG gRNA.
Using flow cytometry, we analyzed the doxycycline response of CRISPRi cells based on mCherry expression (as a surrogate for dCas9-KRAB expression; Figure S5A). There was no silencing of the TetO promoter in low-passage and high-passage iPSCs, suggesting long-term culturing (>3 months) does not cause silencing. However, cardiac progenitors (day 5) and iPS-CM (day 15) lose ~20% and 50–80% of the doxycycline response, respectively. Prolonging the duration of doxycycline treatment (from 2 to 7 days) and splitting the cells improved doxycycline response (as measured by mCherry expression) in iPS-CM (Figure S6C). For this reason, we have initiated all our knockdowns on day 5-post differentiation to obtain the maximum amount of silencing. It is worth noting that with CRISPRi, only minute amounts of the dCas9-KRAB protein is necessary to induce a knockdown. Hence, knockdown might happening even in cells that do not turn on mCherry expression (Figure S5).
The knockdown of the HERG potassium channel in iPSCs was highly efficient (>95%) while the knockdown in iPS-CM was only 60% effective, we hypothesize the reduction in the efficiency of HERG knockdown is partially due to activation of other HERG isoforms in iPS-CM. We further investigated whether knocking down of HERG potassium channel in iPS-CM would recapitulate a physiologically relevant cellular phenotype. We found that knocking down HERG in iPS-CM lead to a prolonged beat duration and the appearance of a shoulder during the down-stroke as measured using the GCaMP signal (which can be used as a surrogate for the action potential) (Huebsch et al., 2015) (Figures 6D and 6E). We confirmed the prolongation of action potential duration by patch-clamp electrophysiology in the HERG knockdown samples (Figures 6F). We expected this result, because the HERG potassium channel pumps K+ ions out of cells to lower the inner membrane potential during diastole. This cellular phenotype recapitulates aspects of the phenotype observed in LQT patients and their iPS-CM (Schwartz et al., 2012; Spencer et al., 2014).
Discussion
In this study, we combined the power of human iPSC technology, which generates functional human cells, with inducible CRISPR-based genome editing and modulation technologies. Using the TetO inducible system we deploy the newly developed CRISPRi system in the AAVS1 safe harbor locus of human iPSC to enable precise control on transcript silencing upon addition of doxycycline. With this approach, we rapidly and efficiently generated loss-of-function phenotypes in iPSCs and their cell-type derivatives to study mechanisms in development and disease. We introduced a single doxycycline-inducible vector system into the AAVS1 safe-harbor locus to gain tight transcriptional control of dCas9-KRAB (for CRISPRi) and Cas9 (for CRISPRn) for gene knockdown and knockout studies, respectively. This inducible vector system helped us precisely control the timing of knocking down the expression of target genes in a clonal iPSC carrying the gRNA of interest. We were also able to efficiently target the CRISPRi vector into non-iPSC human cells (T-lymphocytes) and show efficient levels of transgene knockdown, which demonstrates the versatility of using the CRISPRi system in a wide range of cell types. This system can be readily targeted to other human cellular models in vitro and also to mouse models (Soriano, 1999) by exchanging the AAVS1-homology arms with the ROSA26-specific knock-in arms.
We found that in iPSC populations, CRISPRi produced a homogeneous and rapid loss-of-function phenotype compared to CRISPRn. CRISPRi avoids potential complications associated with incomplete loss-of-function and gain-of-function phenotypes in cell populations produced by Cas9-induced hypomorphic alleles. Therefore, CRISPRi represents a powerful technology for repressing gene expression in bulk populations and especially when performing genome-scale phenotypic screens. Every CRISPRi iPSC that contained a target-specific gRNA displayed a rapid, uniform, and efficient transcriptional knockdown. This result was also validated across multiple endogenous loci in iPSCs, cardiac progenitors and iPS-CM. By contrast, using CRISPRn we found that while all cells harbored the gRNA-expression vector and had continuous expression of Cas9, they did not all display complete loss-of-function phenotypes. Indeed, up to one-third of the cells maintained expression of the target gene. When we sequenced the target alleles, we found that of the mutated alleles, over one-third had in-frame INDELs, potentially resulting in a hypomorphic protein encoded by a gene that is now resistant to further Cas9 cutting using the target gRNA. Statistically, we expect that one-third of the INDELs generated by double-strand breaks induced by Cas9 through the NHEJ pathway would to produce in-frame mutations. This effect could produce partial loss-of-function or gain-of-function phenotypes. Additionally, the location and size of the in-frame INDEL might not change the function of the mutated protein compared with the wild-type protein (Boettcher and McManus, 2015; Shi et al., 2015; Sung et al., 2013).
Since CRISPRi gRNAs were only effective at promoter regions close to the TSS, this may reduce the likelihood of off-target effects by transcriptional interference elsewhere in the genome. Indeed, RNA-Seq analysis showed that the knockdown of GCaMP was highly specific. Furthermore, expression of dCas9-KRAB did not cause significant off-target transcriptional changes as compared to Cas9 expression alone. Although CRISPRi is highly effective, there are cases when other genetic tools such as CRISPRn, TALENs and RNAi may have advantages. For instance, we and others (Gilbert et al., 2014) show that CRISPRi gRNAs are only effective near the TSS, which restricts the efficiency of transcript for genes that have poorly defined or multiple TSSs. While, CRISPRn and TALENs can be effective at any exon as long as the genomic region is accessible (Doench et al., 2014; Kim et al., 2013b). Additionally, RNAi can target any constitutive portion of the mRNA, and has already been approved for human therapy (Davidson and McCray, 2011; Haussecker, 2012), however RNAi has shown to have many off-target effects (Jackson et al., 2003; Kim et al., 2013b; Krueger et al., 2007).
We also demonstrated the feasibility of allele-specific interference and the tunable nature of CRISPRi-based knockdown, which can be used to study the dose-dependent effects of a gene involved in development and disease. The dosage of transcription factors plays a significant role during development and organogenesis (McFadden et al., 2005; Takeuchi et al., 2011). In addition, many human diseases result from haploinsufficiency in which a mutation in a single copy of a gene produces the disease phenotype (Armanios et al., 2005; Marston et al., 2012; Minami et al., 2014; Theodoris et al., 2015). Therefore, to study dosage-dependent effects of transcription factors in development and disease, CRISPRi can be used to homogeneously tune the level of repression in cells by either choosing the relevant gRNA sequences, or empirically titrating the levels of doxycycline to achieve the desired knockdown level. Alternatively, introducing a single point mutation at different positions in the gRNA sequence (which leads to mismatches between the RNA-DNA homology sequence) can be used to tune CRISPRi knockdown activity (Gilbert et al., 2014). Finally, CRISPRi knockdown was reversible in iPSCs upon doxycycline withdrawal, which would support studies involving transient knockdown of transcripts within a specific window during cell differentiation.
Our studies with CRISPRi in iPSCs showed that knocking down transcripts involved in maintaining pluripotency is highly efficient and rapidly causes a complete loss of pluripotent morphology, followed by cell differentiation in all cells expressing the appropriate gRNA. We also used this approach to knockdown the HERG potassium channel to mimic an LQT2-type phenotype in iPS-CM. We found that the inducible TetO promoter is partially silenced during the cardiac differentiation process, and this is reported to be due to methylation at CpG dinucleotides (Oyer et al., 2009). This silencing is independent of integration at the AAVS1 locus, as CAG-driven transgenes integrated at the AAVS1 locus remain active after differentiation. To avoid the effects of promoter silencing we initiated transcript knockdown in the iPSC state or progenitor cells (day 5 of differentiation) where the vast majority of the cells respond to doxycycline. This strategy has proved highly effective at transgene knockdown in cardiac progenitors and iPS-CM. To circumvent issues with silencing in future studies, we generated a non-inducible CRISPRi iPSC line (Gen3; in which dCas9-KRAB is driven off the CAG promoter) and the knockdown can be initiated upon introduction of gRNA. With this cell line, we expect to achieve highly efficient knockdown in differentiated cell types, such as iPS-CM.
Several groups have used the CRISPR/Cas9 system for loss-of-function genetic screens in human cells (Shalem et al., 2013; Wang et al., 2013). Furthermore, some groups have used genome-scale screens with CRISPRi and CRISPR activation (CRISPRa) to identify known and novel genes that control cell growth and sensitivity to cholera-diphtheria toxin (Gilbert et al., 2014). In this study, we present our CRISPRi iPSC lines as suitable model systems for performing screens to identify novel transcripts of pluripotency, drug resistance, and cell survival at the pluripotent stem-cell stage. With genome-scale screens, we can identify factors that improve cell-specific differentiation into functional cell types that have been traditionally hard to obtain, and we can more rapidly generate mature functional cell types that better mimic in vivo cell counterparts. In addition, with CRISPRi, we can repress putative disease-associated genes in a medium-to high-throughput manner to unravel the molecular mechanisms underlying human disease in vitro. Finally, we can build on the current power of CRISPRi for developmental screens by using an orthogonal dCas9-effector system for gene activation via CRISPRa, which can synergistically modulate gene knockdown and activation and direct cell fate towards a particular lineage.
Experimental Procedures
iPSC Culture
WTB and WTC iPSCs and derivative lines were maintained under feeder-free conditions on growth factor-reduced Matrigel (BD Biosciences) and fed daily with mTeSR medium (STEMCELL Technologies) (Ludwig et al., 2006). Accutase (STEMCELL Technologies) was used to enzymatically dissociate iPSCs into single cells. To promote cell survival during enzymatic passaging, cells were passaged with p160-Rho-associated coiled-coil kinase (ROCK) inhibitor Y-27632 (10 µM; Selleckchem) (Watanabe et al., 2007). iPSCs were frozen in 90% fetal bovine serum (HyClone) and 10% dimethyl sulfoxide (Sigma).
Generation of Stable CRISPRi and CRISPRn iPSC Lines
iPSCs were singularized with accutase, resuspended in PBS, and counted with a Countess automated cell counter (Life Technologies). For plasmid transfections, the human stem cell nucleofector kit 1 solution was used on the Amaxa nucleofector 2b device (program A-23; Lonza). To generate the CRISPRi and CRISPRn iPSC lines, 2 million WTC or WTB iPSCs were nucleofected with the appropriate knock-in vector (5 µg) and each AAVS1 TALEN pair (2 µg; Table S1). Cells were then seeded in 6-well plates in serial dilutions in mTeSR supplemented with Y-27632 (10 µM). Selection was applied 3 days post-nucleofection with the appropriate antibiotic in mTeSR plus Y-27632 (10 µM). To knock-in of the CRISPRi construct (carrying the Neomycin resistance gene cassette), Geneticin (Life Technologies) was applied at 100 µg/ml. To knock-in the CRISPRn and GCaMP constructs (carrying the Puromycin resistance-gene cassette), 0.5 µg/ml of Puromycin (Life Technologies) was added. Selection was maintained for ~10 days until stable colonies appeared. Colonies with a diameter of greater than ~500 µm were manually picked using a P200 pipette tip under an EVOS FL picking microscope (Life Technologies) and transferred to individual wells of a 24-well plate containing mTeSR medium supplemented with Y-27632 (10 µM). Clones were then expanded into larger vessel formats.
Generation of CEM CRISPRi Cell Line
CEM CRISPRi cells were generated by electroporation of 0.5 µg of each AAVS1 TALEN pair and 1 µg of the Gen1 CRISPRi vector) with Amaxa nucleofector 2b device and Amaxa cell line nucleofector kit C (Lonza). Cells were selected in 1 µg/µl G418, and clonal lines were generated by dilution in 96-well plates. Clonal populations were selected based on doxycycline induction of mCherry expression. Oligos encoding the CD4 protospacer were annealed and cloned into the pSLQ1371 vector using restriction sites BstXI and BlpI, and lentivirus was produced in HEK293T cells (Gilbert et al., 2014). To compare performance of CD4 gRNAs, each was transduced into CEM-CRISPRi cells. Transduced populations were incubated for 96 h with doxycycline (2 µM). Knockdown efficiency was calculated by gating all mCherry expressing cells, and comparing cell-surface CD4 expression in the presence or absence of gRNA-expressing cells (BFP+). Three independent stable CEM CRISPRi clones were selected with 0.6 µg/mL Puromycin, and incubated in the presence or absence of doxycycline (2 µM) for 14 days to assess maximal CD4 knockdown. Cells were stained using anti-CD4 APC-conjugated antibody and cell surface CD4 staining was quantified using a BD LSRII flow cytometer. CD4 knockdown was quantified as percent reduction relative to no doxycycline treatment condition.
gRNA Design and Cloning into the gRNA-Expression Vector
For CRISPRi, three to five gRNAs were designed to target near the transcription start site (TSS) of the gene of interest (250 bp upstream and downstream, respectively). The location of the TSS was determined using NCBI (http://www.ncbi.nlm.nih.gov/). gRNA oligos were designed, phosphorylated, annealed and cloned into the pgRNA-CKB vector using BsmBI ligation strategy. Additional details and a list of gRNA sequences are listed in supplemental experimental procedures.
gRNA Nucleofection and Selection of Stable CRISPRi and CRISPRn Clones
The gRNA-expression vector (pgRNA-CKB) was transfected into either the CRISPRi or CRISPRn cells with the human stem cell nucleofector kit 1 solution on the Amaxa nucleofector 2b device (program A-23; Lonza). Two million CRISPRi or CRISPRn iPSCs and 5 µg of the circular gRNA-expression plasmid were used per nucleofection. Nucleofected cells were then seeded in a single well of a 6-well plate in mTeSR supplemented with Y-27632 (10 µM). Blasticidin selection (10 µg/ml) was applied 24 h post-nucleofection in mTeSR supplemented with Y-27632 (10 µM) for 7–10 days, until stable colonies appeared. Stable colonies were then pooled and passaged at least three times in mTeSR plus Blasticidin and Y-27632 to enrich for cells with integration at transcriptionally active sites (Figure S3).
RNA Sequencing
For each sample, 1 µg of total RNA was prepared using TRIzol as previously described. Strand-specific mRNAseq libraries were prepared using TruSeq Stranded mRNA Library Prep Kit (Illumina). Upon completion, libraries were quantified and pooled using Qubit dsDNA HS assay, and Agilent’s Bioanalyzer high-sensitivity DNA assay. The indexed libraries were pooled and sequenced on Illumina HiSeq 4000 as 50 bp single-end reads. Reads were aligned to the hg19 genome assembly using the Ensembl 75 reference transcriptome customized to include the GCaMP6f constructs using TopHat2 (Kim et al., 2013a). Unaligned reads were subsequently aligned to the CRISPRi or CRISPRn knockin constructs where appropriate. Transcript alignments were then counted using SubRead v1.4.6 and analyzed with custom scripts written in Python (Liao et al., 2013). All data are displayed as reads per million (RPM) with a pseudocount of 0.075.
iPS-CM Differentiation and Lactate Purification
iPSCs were differentiated into iPS-CM using the WNT modulation-differentiation method (Lian et al., 2012) (Figure S5A). iPS-CM were purified via a modified version of the lactate metabolic-selection method (Tohyama et al., 2013). Additional details are oulined in supplemental experimental procedures.
Supplementary Material
Acknowledgments
We thank members of the Conklin laboratory, Gladstone Institute of Cardiovascular Disease, the Roddenberry Stem Cell Core, BioFulcrum a Gladstone Institutes Enterprise and Innovative Genomics Institute for technical assistance and helpful comments on the manuscript. We thank Tim Rand and Knut Woltjen for valuable discussions and helpful comments on the manuscript. We thank S. John Liu for RNA-seq analysis advice. We thank Jen Berman and Samantha Cooper at Bio-Rad for assistance with designing ddPCR probe-primer sets. Summer students Matthew Keller and Monique Morrison assisted with preliminary experiments. CEM CD4+ cells were obtained from Dr. J.P. Jacobs through the AIDS Reagent Program, Division of AIDS, NIAID, NIH. M.A.M. is supported by the Canadian Institutes of Health Research postdoctoral fellowship 129844. N.H. was supported by the CIRM training program TG2-01160 and T32 HL007544. E.F. was supported by a Bridges to Stem Cell Training grant TB1-01188 from CIRM. L.M.J. is supported by the CIRM Training Grant TG2-01160 and NICHD Career Development Award 1K12HD072222. Y.M. received fellowships from the Uehara Memorial Foundation Research and Gladstone-CIRM. D.G. is supported by UCSF-Gladstone Center for AIDS Research (CFAR), an NIH-funded program (P30 AI027763). JEV, MAH, LAG, JSW were supported by the Howard Hughes Medical Institute and the National Institutes of Health (R01 DA036858). B.R.C. received support from the US National Heart, Lung, and Blood Institute and the National Institutes of Health (U01-HL100406, U01-GM09614, R01-HL108677, U01-HL098179, U01-HL099997, P01-HL089707, R01HL130533 and R01-HL060664).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
M.A.M. and B.R.C. were primarily responsible for conception, design and interpretation of the experiments. M.A.M. conducted most experiments with help from N.H., E.F., E.S., A.T., M.P.O, T.V.E., and L.M.J. Y.M. and A.H.C. generated the CRISPRn Gen1C iPSC line. C.I.S. performed electrophysiology experiments. D.E.G. generated the CEM CRISPRi cell line and provided knockdown analysis. L.A.G., J.S.W. and L.S.Q. provided technical expertise and the CRISPRi fusion cassette and gRNA expression constructs. J.E.V and M.A.H conducted and analyzed the RNA-Seq experiments. M.A.M., P.L.S., and B.R.C. wrote the manuscript with support from all authors.
References
- Apáti Á, Pászty K, Hegedűs L, Kolacsek O, Orbán TI, Erdei Z, Szebényi K, Péntek A, Enyedi Á, Sarkadi B. Characterization of calcium signals in human embryonic stem cells and in their differentiated offspring by a stably integrated calcium indicator protein. Cell. Signal. 2013;25:752–759. doi: 10.1016/j.cellsig.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Armanios M, Chen J-L, Chang Y-PC, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher M, McManus MT. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol. Cell. 2015;58:575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc. Natl. Acad. Sci. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BL, McCray PB. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014 doi: 10.1038/nbt.3026. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F, Zhu Z, Shi Z-D, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D. The Business of RNAi Therapeutics in 2012. Mol. Ther. — Nucleic Acids. 2012;1:e8. doi: 10.1038/mtna.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Caboni L, Das D, Yumoto F, Clayton T, Deller MC, Nguyen P, Farr CL, Chiu H-J, Miller MD, et al. Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells. Proc. Natl. Acad. Sci. 2015;112:4666–4671. doi: 10.1073/pnas.1502855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur A, Nguyen TN, Yoo JC, Judge LM, et al. Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales. Tissue Eng. Part C Methods. 2015;21:467–479. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013a;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013b;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- Krueger U, Bergauer T, Kaufmann B, Wolter I, Pilk S, Heider-Fabian M, Kirch S, Artz-Oppitz C, Isselhorst M, Konrad J. Insights into Effective RNAi Gained from Large-Scale siRNA Validation Screening. Oligonucleotides. 2007;17:237–250. doi: 10.1089/oli.2006.0065. [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. 2012:201200250. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston S, Copeland O, Gehmlich K, Schlossarek S, Carrrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J. Muscle Res. Cell Motil. 2012;33:75–80. doi: 10.1007/s10974-011-9268-3. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Minami SS, Min S-W, Krabbe G, Wang C, Zhou Y, Asgarov R, Li Y, Martens LH, Elia LP, Ward ME, et al. Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med. 2014;20:1157–1164. doi: 10.1038/nm.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, Lizarraga PP, So P-L, Conklin BR. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 2014 doi: 10.1038/nmeth.2840. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyer JA, Chu A, Brar S, Turker MS. Aberrant Epigenetic Silencing Is Triggered by a Transient Reduction in Gene Expression. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz PJ, Crotti L, Insolia R. Long-QT Syndrome From Genetics to Management. Circ. Arrhythm. Electrophysiol. 2012;5:868–877. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2013:1247005. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spencer CI, Baba S, Nakamura K, Hua EA, Sears MAF, Fu C, Zhang J, Balijepalli S, Tomoda K, Hayashi Y, et al. Calcium Transients Closely Reflect Prolonged Action Potentials in iPSC Models of Inherited Cardiac Arrhythmia. Stem Cell Rep. 2014;3:269–281. doi: 10.1016/j.stemcr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterneckert JL, Reinhardt P, Schöler HR. Investigating human disease using stem cell models. Nat. Rev. Genet. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- Sung YH, Baek I-J, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim J-S, Lee H-W. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguín P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoris CV, Li M, White MP, Liu L, He D, Pollard KS, Bruneau BG, Srivastava D. Human Disease Modeling Reveals Integrated Transcriptional and Epigenetic Mechanisms of NOTCH1 Haploinsufficiency. Cell. 2015;160:1072–1086. doi: 10.1016/j.cell.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct Metabolic Flow Enables Large-Scale Purification of Mouse and Human Pluripotent Stem Cell-Derived Cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR/Cas9 System. Science. 2013:1246981. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.