Summary
Small RNAs of 21–25 nucleotides (nt), including small interfering RNAs (siRNAs) and microRNAs (miRNAs), act as guide RNAs to silence target-gene expression in a sequence-specific manner [1]. In addition to a Dicer homolog, DCL1, the biogenesis of miRNAs in Arabidopsis requires another protein, HEN1 [2, 3]. miRNAs are reduced in abundance and increased in size in hen1 mutants [2, 4–7]. We found that HEN1 is a miRNA methyltransferase that adds a methyl group to the 3′-most nucleotide of miRNAs [8], but the role of miRNA methylation was unknown. Here, we show that siRNAs from sense transgenes, hairpin transgenes, and transposons or repeat sequences, as well as a new class of siRNAs known as trans-acting siRNAs, are also methylated in vivo by HEN1. In addition, we show that the size increase of small RNAs in the hen1-1 mutant is due to the addition of one to five U residues to the 3′ ends of the small RNAs. Therefore, a novel uridylation activity targets the 3′ ends of unmethylated miRNAs and siRNAs in hen1 mutants. We conclude that 3′-end methylation is a common step in miRNA and siRNA metabolism and likely protects the 3′ ends of the small RNAs from the uridylation activity.
Results and Discussion
miRNAs in hen1 Mutants Are Heterogeneous in Size
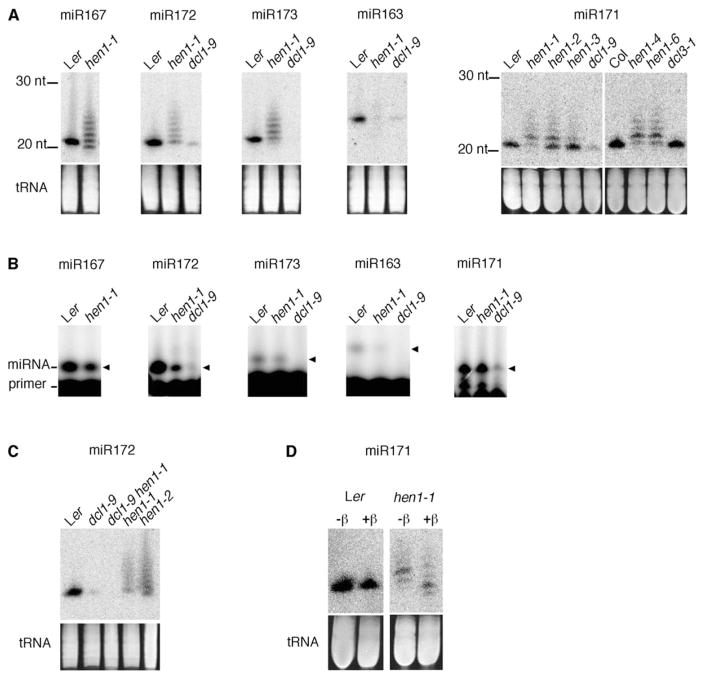
Previous studies showed, with RNA filter hybridization, that miRNAs in hen1 mutants are either reduced in abundance or totally absent [2, 4–7]. For some miRNAs, smearing bands larger than wild-type miRNAs were detected [2, 5]. When we resolved total RNAs from hen1-1 on high-resolution acrylamide gels, heterogeneity in the size of miRNAs was dramatically revealed by the presence of a ladder of bands, and this ladder presumably represents molecules differing in length in 1 nt intervals (Figure 1A). Most of the miRNA species in hen1-1 were larger than those in Ler, ranging from 21 to 26 nt (Figure 1A). miRNA species apparently smaller than wild-type were also detected (Figure 1A, miR163, miR167, and miR173). A similar pattern of miRNA accumulation was observed in other hen1 alleles for miR171 (Figure 1A) and miR172 (Figure 1C). Therefore, HEN1 appears to maintain the correct size as well as the normal levels of miRNAs.
Figure 1. miRNAs in hen1 Have 3′-End Defects.
(A) RNA filter hybridization of miR167 (21 nt), miR172 (21 nt), miR173 (22 nt), miR163 (24 nt), and miR171 (21 nt) in various genetic backgrounds. Ler is the wild-type control for hen1-1, hen1-2, hen1-3, and dcl1-9. Col is the wild-type control for hen1-4, hen1-6 (SALK_090960), and dcl3-1.
(B) 5′ ends of miRNAs in hen1-1 are the same as those in Ler as detected by primer extension. Primer extension was performed as described [24], with 17–18 nt DNA primers spanning the 3′ portions of mature miRNAs. The extension products are indicated by the arrowheads.
(C) Accumulation of miR172 in various genotypes as detected by RNA filter hybridization.
(D) Detection of 3′ methylation of miR171. Total RNAs were treated (+β) or not (−β) with the oxidation and β elimination reagents, and miR171 was detected by RNA filter hybridization. miR171 from the wild-type is resistant, whereas the heterogeneous miR171 species in hen1-1 are sensitive to the chemical reactions. In (A), (C), and (D), images of ethidium-bromide-stained gels in the region of tRNAs are shown at the bottom to indicate near-equal loading.
Heterogeneous miRNA Species in hen1 Are Generated by DCL1
DCL1 is required for the generation of miRNAs [2, 3] and trans-acting siRNAs [9, 10]. The predominant size of these two types of small RNAs is 21–22 nt. DCL3 is required for the accumulation of 24–25 nt endogenous siRNAs but not miRNAs [7]. Loss of DCL3 activity results in a shift in size of siRNA02, an endogenous siRNA, from 24–25 nt to 21–22 nt [7], presumably as a result of the activity of another DCL or other DCLs. To investigate whether the miRNA size heterogeneity in hen1 mutants resulted from the mistargeting of miRNA precursors by DCL proteins other than DCL1, we analyzed the expression of miR172 in the dcl1-9 hen1-1 double mutant (Figure 1C). The absence of miR172 in dcl1-9 hen1-1 indicates that the heterogeneous miR172 species in hen1-1 are all generated by DCL1.
Heterogeneous miRNAs in hen1-1 Have the Same 5′ Ends as Wild-Type miRNAs
To begin to understand how the size heterogeneity of miRNAs in hen1 mutants is generated, we first determined whether the heterogeneity resides in the 5′ ends of the miRNAs by primer extension. Seventeen- to eighteen-nucleotide DNA oligonucleotides complementary to the 3′ portions of miRNAs were used in primer-extension assays on Ler and hen1-1 RNAs. Extension products of the same sizes were obtained in Ler and hen1-1 for all the miRNAs examined (Figure 1B). The specificity of the primer extension was inferred from the reduced signals in the dcl1-9 mutant (Figure 1B), which is known to have decreased or undetectable levels of the miRNAs ([2, 3]; Figure 1A). This indicates that the heterogeneous miRNA species in hen1-1 have the same 5′ ends as the corresponding miRNA species in Ler. Therefore, the size heterogeneity of miRNAs in hen1-1 should reside in the 3′ ends.
miRNAs in hen1-1 Lack 3′ Methylation and Become 3′ Uridylated
How does the 3′-end heterogeneity of miRNAs in hen1 mutants arise? We envisioned that at least two mechanisms account for the 3′-end heterogeneity. First, DCL1 imprecisely processes the pre-miRNAs in the absence of functional HEN1 to generate miRNAs with differing 3′ ends. In this scenario, HEN1 specifies the 3′ ends of the miRNAs in the pre-miRNAs during DCL1 processing. This seems unlikely given HEN1’s role as a miRNA methyltransferase [8]. Second, DCL1 is able to precisely process the pre-miRNAs in hen1 mutants, but additional nucleotides are added to the 3′ ends after miRNAs are generated. This model predicts that the extra nucleotides at the 3′ ends of miRNA species in hen1 are unlikely to correspond to those 3′ to the miRNAs in the pre-miRNAs.
We utilized a modified miRNA-cloning approach to determine the identity of the extra nucleotides in the 3′ ends of miRNAs. Small RNAs of 18–28 nt from Ler and hen1-1 were isolated and ligated to a 3′ adaptor. Two miRNAs (miR167 and miR173) were selectively amplified with miRNA-specific primers (corresponding to the 5′ portion of the miRNAs) and the 3′-adaptor primer. The resulting PCR products were cloned and sequenced as described [2]. In the great majority of wild-type clones (100% for miR173 and 98% for miR167), at least two nucleotides immediately adjacent to the miRNA-specific primer are the same as those in the miRNAs. In most wild-type clones (89% for miR173 and 91% for miR167), at least three nucleotides immediately adjacent to the primer sequence are the same as those in the miRNAs. This indicates that we selectively amplified the miRNAs of interest. Approximately 60% of the clones from the wild-type represent the full-length miRNAs, and the remaining ones are shorter than full length. The shorter miRNA species are either minor species normally present in the wild-type, or they were truncated during the experiment.
As predicted, species longer than miR173 and miR167 at the 3′ ends were cloned in the hen1-1 mutant but not in the wild-type (Table 1). The extra nucleotides did not correspond to the 3′ flanking sequences in their precursors. For example, among the cloned miR173 species that were full length or longer in the hen1-1 mutant (H5 to H10), 38% (H6 to H10) had extra nucleotides. Most of the extra nucleotides were U residues instead of those 3′ to miR173 in the precursor. For miR167, one clone (H13) had U residues after the full-length miRNA sequence. Most cloned miR167 species from hen1-1 (H1 to H11) were not full-length miR167. It is very unlikely that they represent degradation intermediates from the handling of the RNAs in the experiment because many had U tails. It is unlikely that these U residues were artificially incorporated during our cloning procedure or that miRNAs with 3′ U residues were preferentially cloned because our previous miRNA cloning with a similar RNA-ligase based approach did not reveal a preference for miRNAs with 3′ U residues [2]. The miR173 and miR167 species cloned from the wild-type also did not reveal a preference for 3′ U residues. Therefore, we believe that the 3′-truncated miR167 species with U tails represent miR167 species that are present in vivo. Occasionally, other nucleotides were also found in the tails of miRNAs from hen1-1 (Table 1).
Table 1.
miR167 and miR173 Cloned from Ler and hen1-1
| Name | Sequencesa | Length (nt) | Number of Clones | Genotype |
|---|---|---|---|---|
| Sequences from the miR173 Cloning | ||||
|
| ||||
| miR173 | UUCGCUUGCAGAGAGAAAUCACagugguca | 22 | Ler | |
| L1 | UUCGCUUGCAGAGAGAAAUC | 20 | 2 | Ler |
| L2 | UUCGCUUGCAGAGAGAAAUCu | 21 | 2 | Ler |
| L3 | UUCGCUUGCAGAGAGAAAUCA | 21 | 9 | Ler |
| L4 | UUCGCUUGCAGAGAGAAAUCAa | 22 | 2 | Ler |
| L5 | UUCGCUUGCAGAGAGAAAUCAC | 22 | 21 | Ler |
| H1 | UUCGCUUGCAGAGAGAAAUau | 21 | 1 | hen1-1 |
| H2 | UUCGCUUGCAGAGAGAAAUCc | 21 | 1 | hen1-1 |
| H3 | UUCGCUUGCAGAGAGAAAUCA | 21 | 2 | hen1-1 |
| H4 | UUCGCUUGCAGAGAGAAAUCAu | 22 | 1 | hen1-1 |
| H5 | UUCGCUUGCAGAGAGAAAUCAC | 22 | 21 | hen1-1 |
| H6 | UUCGCUUGCAGAGAGAAAUCACa | 23 | 1 | hen1-1 |
| H7 | UUCGCUUGCAGAGAGAAAUCACu | 23 | 7 | hen1-1 |
| H8 | UUCGCUUGCAGAGAGAAAUCACuu | 24 | 2 | hen1-1 |
| H9 | UUCGCUUGCAGAGAGAAAUCACuuc | 25 | 1 | hen1-1 |
| H10 | UUCGCUUGCAGAGAGAAAUCACuuu | 25 | 2 | hen1-1 |
|
| ||||
| Sequences from the miR167 Cloning | ||||
|
| ||||
| miR167a | UGAAGCUGCCAGCAUGAUCUAauuagcuu | 21 | Ler | |
| miR167b | UGAAGCUGCCAGCAUGAUCUAucuuuggu | 21 | Ler | |
| miR167c | UGAAGCUGCCAGCAUGAUCUUgucuuccu | 21 | Ler | |
| miR167d | UGAAGCUGCCAGCAUGAUCUGGuaaucg | 22 | Ler | |
| L1 | UGAAGCUGCCAGCAUGAUu | 19 | 1 | Ler |
| L2 | UGAAGCUGCCAGCAUGAUC | 19 | 2 | Ler |
| L3 | UGAAGCUGCCAGCAUGAUCaa | 21 | 1 | Ler |
| L4 | UGAAGCUGCCAGCAUGAUCU | 20 | 13 | Ler |
| L5 | UGAAGCUGCCAGCAUGAUCUA | 21 | 26 | Ler |
| L6 | UGAAGCUGCCAGCAUGAUCUGG | 22 | 1 | Ler |
| H1 | UGAAGCUGCCAGCAUGAUu | 20 | 1 | hen1-1 |
| H2 | UGAAGCUGCCAGCAUGAUuu | 22 | 2 | hen1-1 |
| H3 | UGAAGCUGCCAGCAUGAUuuuu | 24 | 1 | hen1-1 |
| H4 | UGAAGCUGCCAGCAUGAUuuuuu | 25 | 2 | hen1-1 |
| H5 | UGAAGCUGCCAGCAUGAUC | 20 | 3 | hen1-1 |
| H6 | UGAAGCUGCCAGCAUGAUCa | 21 | 1 | hen1-1 |
| H7 | UGAAGCUGCCAGCAUGAUCU | 21 | 8 | hen1-1 |
| H8 | UGAAGCUGCCAGCAUGAUCUub | 22 | 4 | hen1-1 |
| H9 | UGAAGCUGCCAGCAUGAUCUuub | 23 | 4 | hen1-1 |
| H10 | UGAAGCUGCCAGCAUGAUCUuuub | 24 | 1 | hen1-1 |
| H11 | UGAAGCUGCCAGCAUGAUCUuuuub | 25 | 1 | hen1-1 |
| H12 | UGAAGCUGCCAGCAUGAUCUA | 22 | 2 | hen1-1 |
| H13 | UGAAGCUGCCAGCAUGAUCUAuuu | 25 | 1 | hen1-1 |
Sequences of the miR173 and miR167 genomic loci and the cloned sequences from Ler (L) and hen1-1 (H) are shown. The nucleotides in mature miRNAs are in uppercase letters. The 3′ flanking sequences in the genomic loci are represented by italic, lowercase letters. The sequence of the 5′ cloning primer used for PCR is underlined. Nucleotides in the cloned sequences that do not correspond to those in the mature miRNAs are in lowercase letters. For miR167, the sequences from four different loci (a, b, c, and d) in the genome (http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml) are listed individually; these loci potentially encode three forms of miR167. The expression of each individual has not been examined previously.
These sequences can theoretically be regarded as miR167c and miR167c with U tails. However, because miR167c was never cloned from Ler, we believe they are truncated miR167a, b, or d with U tails.
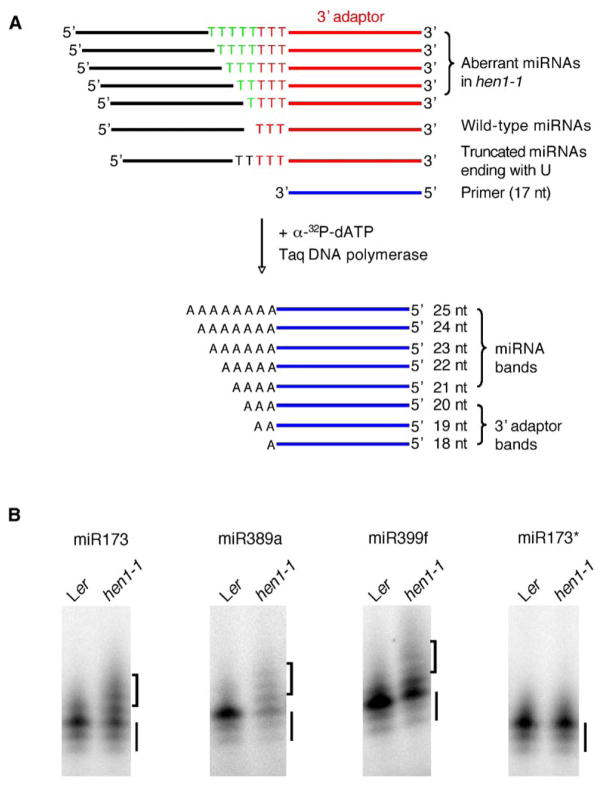
To further confirm the presence of U tails in these and other miRNAs, we carried out a primer-extension assay on the previously mentioned miRNA-specific RT-PCR products amplified from miRNAs ligated to adaptors. Taq DNA polymerase was used to extend the RT-PCR products with a primer complementary to the 3′ portion of the 3′ adaptor, in the presence of only α-32P-dATP (Figure 2A). As a result of the presence of three T residues at the 5′ end of the adaptor, products terminating at each of the T residues were generated in both wild-type and hen1-1 samples (bars in Figure 2B). Whereas further extension beyond the adaptor was absent or rare in wild-type, a ladder of products with A residues was obvious in hen1-1 (brackets in Figure 2B). To rule out the remote possibility of other deoxynucleotides contaminating the RT-PCR products that were purified from polyacrylamide gels, we also performed the primer extension with a primer complementary to the entire 3′-adaptor sequence in the presence of any one of the four radioactive deoxynucleotides. A ladder of primer-extension products was only observed in the hen1-1 mutant when dATP, but not any of the other deoxynucleotides, was present (see Figure S1 in the Supplemental Data available with this article online). This confirms that the 3′ ends of miRNAs have U tails in hen1-1. We also carried out the assay on two miRNAs, miR389a1 and miR399f, whose production is DCL1 independent but requires HEN1 [11], and we obtained similar results (Figure 2B). We amplified by RT-PCR the putative pre-miR167a [3] from hen1-1 and confirmed by sequencing that extra U residues were not present in the precursor (data not shown). Therefore, we conclude that the U residues are added after DCL1-mediated cleavage of the pre-miRNAs and that a novel enzymatic activity, presumably that of a terminal transferase or polymerase, adds the U residues to the 3′ ends of miRNAs in hen1-1.
Figure 2. miRNAs in hen1-1 Are Uridylated at Their 3′ Ends.
(A) A schematic diagram of an α-32P-dATP incorporation assay. The first three residues in the 3′ adaptor are T (red). T residues between the mature miRNAs (black line) and the 3′ adaptor (red line and letters) in the RT-PCR products are in green. The primer complementary to the 3′ adaptor (excluding the three T residues) is in blue.
(B) α-32P-dATP incorporation assay performed on miR173, miR389a1, miR399f, and miR173* from Ler and hen1-1. The bottom three bands marked by the bars represent the extension products corresponding to the Ts in the 3′ adaptor. Extension products marked by the brackets indicate the T residues between the mature miRNAs and the 3′ adaptor.
Recently, HEN1 was found to be a miRNA methyltransferase that adds a methyl group to the ribose of the last nucleotide of miRNAs [8]. Whereas miRNAs in the wild-type have a methyl group, the heterogeneous species in hen1-1 lack methylation [8]. The methylation status of small RNAs can be evaluated by treatment of the RNAs with sodium periodate (oxidation) followed by β elimination [12]. RNAs with free 2′ and 3′ OH are sensitive to the chemical-modification reactions, which result in the elimination of the last nucleotide. The resulting RNAs would migrate faster in gel electrophoresis by approximately 2 nt as a result of the presence of a 3′ P [8, 12, 13]. We found that all miR171 species in hen1-1 changed their apparent mobility after the chemical treatment, whereas miR171 from wild-type did not (Figure 1D). Similar analyses in different hen1 alleles revealed that the state of 3′ methylation was inversely correlated with the accumulation of 3′-uridylated miRNAs. For example, the majority of miR171 in hen1-3, a weak allele, was still methylated (data not shown) and appeared wild-type in size (Figure 1A), whereas almost all miR171 species in the severe hen1-1 allele lacked methylation (Figure 1D) and accumulated as 3′-uridylated species (Figure 1A). This suggests that methylation prevents or attenuates the uridylation activity.
The 3′ Ends of the Two Strands of the miRNA/miRNA* Duplex Are Asymmetrically Affected in hen1
Experimental evidence supports the existence of the predicted intermediate, miRNA/miRNA* duplex, in miRNA biogenesis in plants [3, 14, 15]. The sense strand is selectively assembled into an RNA-induced silencing complex (RISC), whereas the antisense strand (miRNA*) is presumably degraded [16–18]. We analyzed the 3′-end sequences of miR173* in the wild-type and hen1-1 by a similar directional-cloning approach described above. Only 1 out of 18 clones in the miR173* population from hen1-1 had extra Us at the 3′ end (Table 2), a proportion that is much lower than the 33.3% (13/39) in the miR173 population (Table 1). We further confirmed this observation by using the α-32P-dATP incorporation assay on the RT-PCR products of miR173* ligated to the 3′ linker. This assay failed to detect 3′-uridylated miR173* in either the wild-type or hen1-1 (Figure 2B). Therefore, there is a strand bias for 3′ uridylation similar to that for RISC assembly [17, 18]. Perhaps, the U tailing enzyme favors the miRNA over miRNA* in the miRNA/miRNA* duplex. It is also possible that this activity acts at a stage after RISC assembly. miR173* clones ending with a nonencoded A residue were also obtained in both wild-type and hen1-1 samples (L2 and H1 in Table 2). It remains to be determined whether they represent in vivo miR173* species or merely nonspecific amplification products.
Table 2.
miR173* Cloned from Ler and hen1-1
| Name | Sequencesa | Length (nt) | Number of Clones | Genotype |
|---|---|---|---|---|
| miR173* | GAUUCUCUGUGUAAGCGAAAGagcuug | 21 | Ler | |
| L1 | GAUUCUCUGUGUAAGCGAAA | 20 | 2 | Ler |
| L2 | GAUUCUCUGUGUAAGCGAAAa | 21 | 1 | Ler |
| L3 | GAUUCUCUGUGUAAGCGAAAG | 21 | 14 | Ler |
| H1 | GAUUCUCUGUGUAAGCGAAAa | 21 | 3 | hen1-1 |
| H2 | GAUUCUCUGUGUAAGCGAAAG | 21 | 14 | hen1-1 |
| H3 | GAUUCUCUGUGUAAGCGAAAGu | 22 | 1 | hen1-1 |
The sequences of the miR173* genomic locus and cloned miR173* sequences from Ler (L1–L3) and hen1-1 (H1–H3) are shown. The nucleotides in mature miR173* are in uppercase letters. The 3′ flanking sequence in the genomic locus is represented by italic, lowercase letters. The sequence of the 5′ cloning primer used for PCR is underlined. Nucleotides that are in the cloned sequences and do not correspond to those in the mature miRNAs are in lowercase letters.
A 24 nt Endogenous siRNA Lacks 3′ Methylation and Becomes 3′ Uridylated in hen1
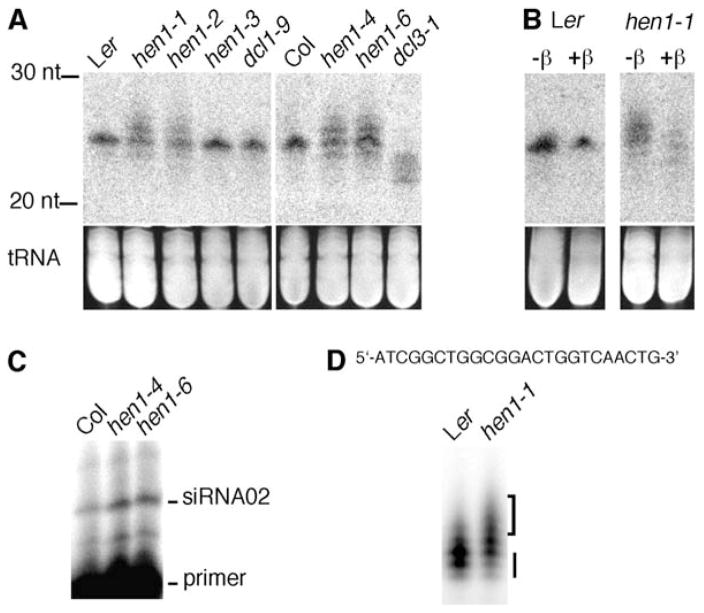
HEN1 is required for the accumulation of some 24–25 nt endogenous siRNAs [7, 19]. Whereas AtSN1 siRNAs and siRNA1003 are barely detectable in hen1-1, the abundance of siRNA02 and cluster2 siRNAs is increased in hen1-1 [7]. Intriguingly, the latter two siRNAs also appear larger in hen1-1 than in the wild-type [7], suggesting that endogenous siRNAs may undergo 3′ uridylation similar to that of miRNAs.
We found that siRNA02 in all the hen1 alleles except the weak hen1-3 allele appeared as a ladder of molecules 23–26 nt in length, in contrast to the 24 nt siRNA02 species in the wild-type (Figure 3). Primer extension of siRNA02 in hen1-4 and hen1-6 showed that the 5′ ends of siRNA02 in these hen1 alleles were the same as that in the wild-type (Figure 3C). Therefore, the heterogeneity in size of this endogenous siRNA resides in the 3′ ends. By using the α-32P-dATP incorporation assay, we further demonstrated that these extra nucleotides are composed of several U residues (Figure 3D). We treated total RNAs with periodate followed by β elimination and detected siRNA02 by RNA filter hybridization to determine the methylation status of this siRNA. Indeed, the apparent mobility of siRNA02 after the reactions remained unchanged in the wild-type but appeared faster in hen1-1 (Figure 3B). We conclude that siRNA02 is methylated by HEN1 in vivo.
Figure 3. A 24 nt Endogenous siRNA in hen1 Is Unmethylated and 3′ Uridylated.
(A) Accumulation of siRNA02 in various genotypes as detected by RNA filter hybridization.
(B) Detection of 3′ methylation of siRNA02. Total RNAs were treated (+β) or not (−β) with the oxidation and β elimination reagents, and siRNA02 was detected by filter hybridization. siRNA02 is methylated in Ler and unmethylated in hen1-1. Images of ethidium-bromide-stained gels in the region of tRNAs in (A) and (B) are shown at the bottom to indicate near-equal loading.
(C) The 5′ ends of siRNA02 in hen1-4 and hen1-6 are the same as those in the wild-type as detected by primer extension. A 15 nt DNA primer complementary to the 3′ portion of siRNA02 was used.
(D) U tails at the 3′ ends of siRNA02 in hen1-1 were detected by an α-32P-dATP incorporation assay (diagrammed in Figure 2A). The bottom three bands marked by the bars represent the extension products corresponding to the three Ts in the 3′ adaptor. Extension products marked by the brackets indicate the T residues between the mature siRNA and the 3′ adaptor. The sequence of siRNA02 is shown. The presence of an extension product 1 nt beyond the 3′ adaptor in the wild-type is probably due to the presence of some siRNA02 species ending with the penultimate T nucleotide.
S-PTGS, IR-PTGS, and trans-Acting siRNAs Are Methylated by HEN1
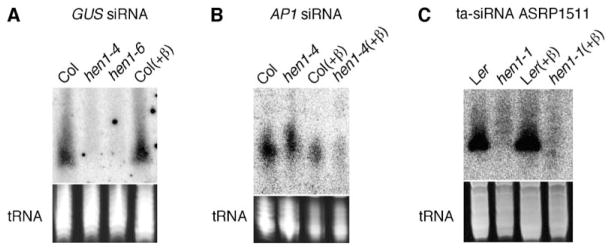
HEN1 was previously found to be required for S-PTGS and for the accumulation of siRNAs from sense transgenes [4]. GUS siRNAs from the silenced L1 locus in the wild-type were resistant to the oxidation and β elimination reactions, suggesting that they are methylated at the last nucleotide (Figure 4A). Although the greatly reduced accumulation of GUS siRNAs in hen1-4 and hen1-6 made it difficult to evaluate whether the siRNAs lack methylation in the hen1 mutants, methylation is likely conferred by HEN1 given its role in S-PTGS.
Figure 4. siRNAs Produced from Transgenes and a ta-siRNA Are Methylated in the Wild-Type but Not in hen1.
(A) Accumulation of S-PTGS siRNAs from GUS transgene (L1 locus) in the wild-type (Col) before and after the oxidation and β elimination reactions (+β) and in hen1-4 and hen1-6 (SALK_090960).
(B) Accumulation of IR-PTGS siRNAs from an AP1 hairpin transgene in the wild-type (Col) and hen1-4 before and after (+β) the oxidation and β elimination reactions.
(C) Accumulation of a ta-siRNA in the wild-type (Ler) and hen1-1 before and after (+β) the oxidation and β elimination reactions. Images of ethidium-bromide-stained gels in the region of tRNAs are shown at the bottom.
The accumulation and function of IR-PTGS siRNAs are not affected by mutations in HEN1 [4]. Our observation that both wild-type and hen1-4 plants carrying an AP1 IR-PTGS construct [20] show ap1 loss-of-function phenotypes (data not shown) supports the notion that HEN1 is not required for IR-PTGS. However, siRNAs from the AP1 hairpin transgene were resistant to the oxidation and β elimination reactions in the wild-type but were sensitive to the reactions in the hen1-4 mutant, suggesting that IR-PTGS siRNAs are also methylated by HEN1 (Figure 4). Moreover, IR-PTGS AP1 siRNAs appeared larger in hen1-4 than in the wild-type (Figure 4B), suggesting that they may also be targeted by the uridylation activity in the absence of methylation. IR-PTGS and S-PTGS are thought to involve overlapping but also distinct pathways because a number of genes (including HEN1) required for S-PTGS are genetically dispensable for IR-PTGS [4, 21]. Our finding that IR-PTGS siRNAs are subjected to methylation by HEN1 as are S-PTGS siRNAs shows that differential genetic requirements do not preclude similar underlying biochemical pathways in the biogenesis and function of S-PTGS and IR-PTGS siRNAs. Perhaps the high levels of IR-PTGS siRNAs allow a bypass of some of the genetic requirements for S-PTGS.
HEN1 is required for the accumulation of trans-acting siRNAs (ta-siRNAs) [9, 10]. However, this requirement may be explained by the fact that the biogenesis of ta-siRNAs depends on certain miRNAs [22] whose biogenesis requires HEN1. However, a direct role of HEN1 in methylating the ta-siRNAs cannot be excluded. We found that ASRP1511, a ta-siRNA [22], was resistant to the chemical-modification reactions in wild-type by not in hen1-1 plants (Figure 4C), indicating that this ta-siRNA is methylated in vivo by HEN1. In addition, the size of this ta-siRNA was heterogeneous in hen1-1 (Figure 4C), suggesting that it is also U tailed in the absence of methylation.
We conclude that methylation on the ribose of the last nucleotide by HEN1 is a universal step in the biogenesis of miRNAs and siRNAs in plants. At least one function of the methylation is to prevent a novel uridylation activity from targeting the last nucleotide of the small RNAs. It was found that U residues are added to the 5′ cleavage products of miRNA-guided cleavage of mRNAs and that the U tailing correlates with the exonucleolytic degradation of the 5′ cleavage products [23]. Perhaps the same enzyme targets unmethylated small RNAs for uridylation, which may subsequently lead to the degradation of the small RNAs. In addition to the uridylation activity, a 3′-to-5′ exonuclease activity is apparently counteracted by the 3′ methylation as suggested by the presence of 3′-truncated miRNAs and siRNAs in hen1 (Figure 1; Table 1). The methylation of miRNAs and siRNAs by HEN1 may also prevent RNA-dependent RNA polymerases (RdRPs) from using the small RNAs as primers. However, the effect of the methyl group on the ability of RdRPs to use the small RNAs as primers needs to be experimentally evaluated.
Experimental Procedures
Directional Cloning and Sequencing of Specific miRNAs
microRNA cloning and sequencing were performed as described [2] with some modifications. In brief, 18–28 nt small RNAs were recovered from total RNAs after gel electrophoresis, dephosphorylated, and ligated to a 3′ adaptor. The ligation products were reverse transcribed with a primer targeting the 3′ adaptor. The RT products were PCR-amplified with a primer corresponding to the 3′ adaptor and ones corresponding to the 5′ portions of specific miRNAs. The PCR products were concatamerized, cloned, and sequenced.
α-32P-dATP Incorporation Assay
Small-RNA purification, 3′-adaptor ligation, and RT- PCR were first performed as described above. miRNA RT-PCR products were PAGE-purified to remove the free nucleotides. The incorporation reactions were run against the purified RT-PCR products with a primer complementary to the 3′-adaptor primer at 94°C for 30 s, 55°C for 30 s, and 72°C for 10 s in the presence of α-32P-dATP and Taq DNA polymerase. The final products were resolved in 15% denaturing PAGE gels and analyzed with a Storm PhosphorImager.
Acknowledgments
We thank Dr. Hervé Vaucheret for providing hen1-4, sgs2-1 (L1), and hen1-4 (IR-PTGS AP1) seeds; Falshruti Patel for assistance in seed planting, plant-genomic-DNA isolation and plant care; and Dr. Sizolwenkosi Mlotshwa for advice on transgene siRNA detection. We thank Drs. Randall Kerstetter, Megerditch Kiledjian, Terry Kinzy, Guiliang Tang, Nilgun Tumer, and Philip Zamore for stimulating discussions. We thank Drs. Randy Kerstetter, Sizolwenkosi Mlotshwa, and Li Zhao for comments on the manuscript. We acknowledge the SALK Institute Genomic Analysis Laboratory (SIGnAL) and the Arabidopsis Biological Resource Center (ABRC) for the generation and distribution of T-DNA insertion lines. This work was supported by a National Sciences Foundation grant (MCB-0343480) to X.C. and a Benedict-Michael Graduate Fellowship from Rutgers University to J.L.
Footnotes
Supplemental Data
Supplemental Data include one supplemental figure and are available with this article online at http://www.current-biology.com/cgi/content/full/15/16/1501/DC1/.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutet S, Vazquez F, Liu J, Beclin C, Fagard M, Gratias A, Morel JB, Crete P, Chen X, Vaucheret H. Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for micro-RNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 10.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alefelder S, Patel BK, Eckstein F. Incorporation of terminal phosphorothioates into oligonucleotides. Nucleic Acids Res. 1998;26:4983–4988. doi: 10.1093/nar/26.21.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutvágner G, McLachlan J, Pasquinelli AE, Balint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 14.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 18.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, He Y, Amasino R, Chen X. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 2004;18:2873–2878. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 22.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. Micro-RNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]