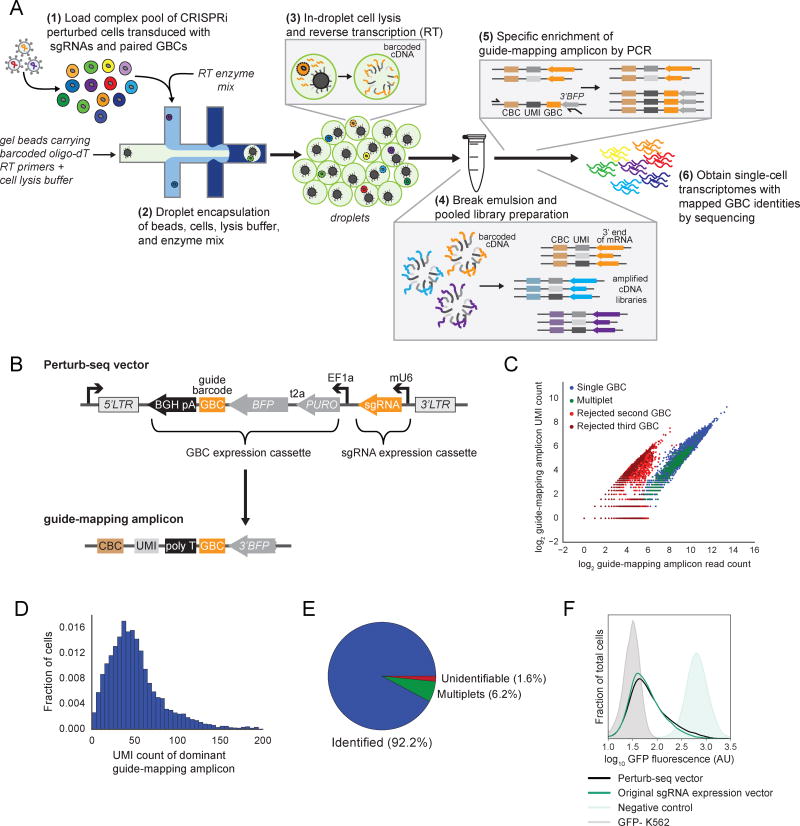
Figure 1. A robust strategy for genetic screens using single-cell gene expression profiling.
(A) Schematic of the Perturb-seq platform. CBC, cell barcode (index unique to each bead). UMI, unique molecular identifier (index unique to each bead oligo). GBC, guide barcode (index unique to each sgRNA).
(B) Schematic of the Perturb-seq vector and guide-mapping amplicon.
(C) Performance of GBC capture. Top 3 possible GBCs for each CBC. CBC identity was assigned to sgRNA identity when a single GBC dominated (blue dots) and any lower abundance GBCs were rejected (red dots). CBC was identified as a “multiplet” when a second or third GBC also had good coverage (green dots). Compare with (D,E).
(D) Distribution of captured UMIs from dominant guide-mapping amplicons.
(E) Performance of perturbation (sgRNA) identification. Data also represented in Figure S1B.
(F) Kernel density estimates of normalized flow cytometry counts representing GFP expression and knockdown achieved from the indicated sgRNA expression constructs.
See also Figure S1.