Abstract
The extent to which non-coding mutations contribute to Mendelian disease is a major unknown in human genetics. Relatedly, the vast majority of candidate regulatory elements have yet to be functionally validated. Here, we describe a CRISPR-based system that uses pairs of guide RNAs (gRNAs) to program thousands of kilobase-scale deletions that deeply scan across a targeted region in a tiling fashion (“ScanDel”). We applied ScanDel to HPRT1, the housekeeping gene underlying Lesch-Nyhan syndrome, an X-linked recessive disorder. Altogether, we programmed 4,342 overlapping 1 and 2 kb deletions that tiled 206 kb centered on HPRT1 (including 87 kb upstream and 79 kb downstream) with median 27-fold redundancy per base. We functionally assayed programmed deletions in parallel by selecting for loss of HPRT function with 6-thioguanine. As expected, sequencing gRNA pairs before and after selection confirmed that all HPRT1 exons are needed. However, HPRT1 function was robust to deletion of any intergenic or deeply intronic non-coding region, indicating that proximal regulatory sequences are sufficient for HPRT1 expression. Although our screen did identify the disruption of exon-proximal non-coding sequences (e.g., the promoter) as functionally consequential, long-read sequencing revealed that this signal was driven by rare, imprecise deletions that extended into exons. Our results suggest that no singular distal regulatory element is required for HPRT1 expression and that distal mutations are unlikely to contribute substantially to Lesch-Nyhan syndrome burden. Further application of ScanDel could shed light on the role of regulatory mutations in disease at other loci while also facilitating a deeper understanding of endogenous gene regulation.
Keywords: non-coding, CRISPR screen, deletion, genome editing, gene regulation, Mendelian, Lesch-Nyhan, paired guide RNA, tiling, ScanDel
Introduction
The success of human genetics in identifying the genes and mutations underlying Mendelian diseases has been facilitated by the incontrovertible reality that the majority of causal mutations lie in protein-coding sequences or splice junctions. Indeed, this assumption is explicit in both classic and contemporary practices in genetics (e.g., exome sequencing). However, it is clear that distal non-coding mutations make some contribution to Mendelian disease. Understanding how often non-coding mutations play a causal role and developing best practices for pinpointing those that do are critical challenges for the field. For example, in the clinic, even if a person is diagnosed with a monogenic Mendelian disorder on the basis of phenotype, clinical sequencing mainly of coding regions fails to identify a causal mutation ∼10% of the time.1 However, possible explanations include not only distal regulatory mutations but also misdiagnosis, somatic mutation, technical false negatives, and others. Furthermore, non-coding loci could contribute to the estimated ∼25%–50% of undiagnosed but apparently Mendelian cases in which the underlying gene is unknown.1, 2
The picture is very different for the genetics of common disease, where over 90% of disease-associated SNPs fall in non-coding regions.3 Many resources have been developed to predict the location of putative regulatory elements and the effects of regulatory mutations,4, 5, 6 such that ∼88% of all protein-coding genes are tied to a cis-expression quantitative locus (eQTL),7 ∼80% of the genome is annotated with biochemical function,8 and numerous tools link regulatory elements to their target genes.9, 10, 11, 12 However, the vast majority of these predictions either are confounded (e.g., by linkage disequilibrium for cis-eQTLs) or lack functional validation. Indeed, there are few distal non-coding regulatory elements that we can confidently assign to a target gene or for which we understand the consequences of disruption.
Large-scale functional experiments are clearly an important next step for the genetics of both common diseases (to facilitate the identification of causal regulatory variants and their target genes) and rare diseases (to identify distal regulatory elements for Mendelian-disease-related genes where causal non-coding mutations might be found). A number of important studies have undertaken functional work to identify and characterize causal or risk-contributory non-coding variants for specific rare and common diseases (e.g., Wakabayashi et al.,13 Weedon et al.,14 and Claussnitzer et al.15) but by approaches that are not easily scalable. Within the last year, several studies have used CRISPR/Cas9 genome editing in cell-based screens to introduce and functionally assay large numbers of non-coding mutations at an unprecedented scale.16, 17, 18, 19, 20, 21 The common approach of these studies is to introduce complex libraries of guide RNAs (gRNAs) via lentiviral infection to a population of cells at a low multiplicity of infection (MOI) and then run an assay that queries the function or expression of a gene of interest. CRISPR/Cas9 mediates double-stranded breaks at sites specified by the gRNA in each cell, eventually resulting in a mutation at each targeted site via imperfect non-homologous end joining (NHEJ).
A fundamental limitation of these singleton-gRNA screens is that because of design constraints (e.g., the uneven distribution of protospacer adjacent motif [PAM] sequences, the variable efficiency of gRNAs, and others), the resulting coverage of regions of interest is incomplete and uneven. Because the majority of bases will be perturbed by zero or only one gRNA, these studies rely on the aggregate behavior of clusters of target sites within potential regulatory elements16 or arbitrarily sized windows (e.g., 500 bp)21 rather than redundant targeting of each base pair by independent gRNAs. Furthermore, it is possible that the mutations introduced by NHEJ at single sites (highly heterogeneous but mainly dominated by small 1–10 bp deletions22, 23) are insufficient to fully disrupt many regulatory elements. Several recent studies have employed an inhibitory domain guided by nuclease-inactive Cas9 to screen non-coding regulatory regions, i.e., CRISPRi.24, 25 Epigenetic modifications mediated by these domains can spread to regions on the order of ∼200 bp to 4.5 kb26, 27 and thus mitigate the challenges related to redundancy and coverage of individual-gRNA screens. However, CRISPRi screens could be less precise because of this spreading effect and, furthermore, do not directly test the consequences of alterations in primary sequence.
Here, we sought to overcome these weaknesses by introducing pairs of gRNAs to each cell with the goal of inducing a kilobase-scale deletion of the intervening DNA between two programmed cuts. A principal advantage of this method (scanning deletion or “ScanDel”) is that tiling deletions across a region allows each targeted base pair to be covered with high redundancy. Furthermore, kilobase-scale deletions are much more likely to eliminate the function of an overlapping or fully contained regulatory element than are small indels resulting from NHEJ at a single target site. Our approach is analogous to classic deletion-scanning experiments28, 29 but has advantages in throughput and in targeting much larger regions in the endogenous genome rather than sequences cloned to a plasmid. Similar strategies have recently been described for the interrogation of long non-coding RNA (lncRNA) genes30 and non-coding sequences.31 Critically, these implementations (and indeed, all CRISPR genetic screens) rely on indirectly genotyping the lentivirally inserted gRNA sequences instead of using direct sequencing of edited loci to confirm exactly which CRISPR-induced genotypes are driving effects.
Here, we applied ScanDel to survey the genomic locus encompassing HPRT1 (MIM: 308000), which encodes the enzyme hypoxanthine(-guanine) phosphoribosyltransferase (HPRT). HPRT1 is a housekeeping gene, a class of genes primarily defined by their broad expression and for which the underlying regulatory architecture remains unclear.32 Loss-of-function mutations in HPRT1 result in X-linked Lesch-Nyhan syndrome33 (MIM: 300322), in which a minority of individuals present with reduced HPRT enzymatic activity despite the absence of identifiable coding mutations.34 Such individuals could carry non-coding mutations that result in reduced HPRT1 expression. Reduced HPRT activity also causes resistance to the drug 6-thioguanine (6TG), a purine analog and chemotherapeutic agent. Thus, it is straightforward to assay cell populations for loss of HPRT1 function, given that only cells with highly reduced expression of functional HPRT will survive selection by 6TG (Figure 1C). Although there are no known distal regulatory elements of HPRT1, its nine exons serve as internal controls.
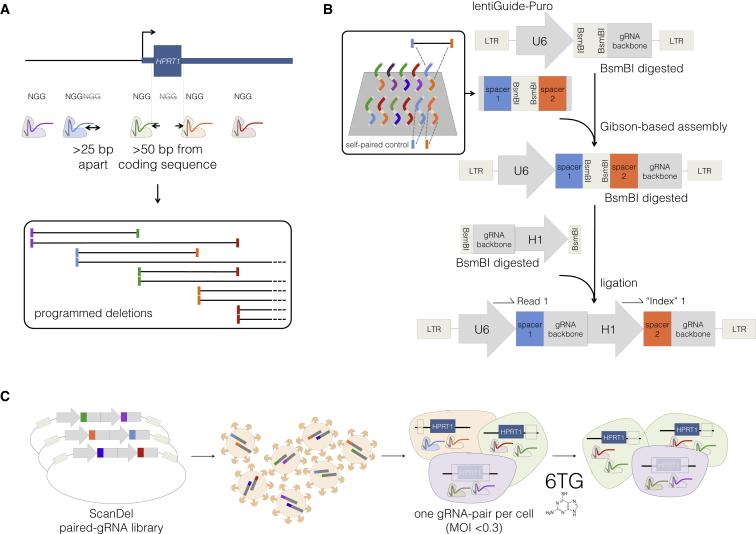
Figure 1.
Design, Delivery, and Selection of ScanDel Library of CRISPR/Cas9-Programmed Deletions for Identification of Non-coding Regulatory Elements
(A) gRNA pairs were designed from a filtered set of protospacers from all Cas9 PAM sequences (5′-NGGs) in the HPRT1 locus (see also Figure 2A). Sites that were >25 bp apart or >50 bp away from exons were kept. For tile design, each remaining spacer was paired to two downstream spacers targeting sequence ∼1 and ∼2 kb away. This resulted in high redundancy of independently programmed, overlapping deletions across the locus (see also Figure 2B).
(B) All spacer pairs corresponding to programmed deletions were synthesized on a microarray (inset). Each spacer was also synthesized as a self-pair as a control for its independent effects. If a self-paired spacer scored positively in the screen, any pairs that used that spacer were removed from the analysis (Figure S1). U6 and gRNA backbone sequence flanked the spacer pairs for Gibson-mediated cloning into lentiGuide-Puro,35 and mirrored BsmBI cut sites separated the spacer pairs to facilitate insertion of a second gRNA backbone and the H1 promoter. In the final library, each gRNA was expressed from its own PolIII promoter. This design facilitates PCR and direct sequencing-based quantification of gRNA-pair abundances.
(C) The lentiviral library of gRNA pairs was cloned at a minimum of 20× coverage (in relation to library complexity) and transduced into HAP1 cells stably expressing Cas9 (via lentiCas9-Blast35) at low MOI. After a week of puromycin selection, the cells were sampled for measurement of the baseline abundance of each gRNA pair. The final cell population was harvested after a week of 6-thioguanine (6TG) treatment, which selected for cells that had lost HPRT enzymatic function. The phenotypic prevalence of each programmed deletion was quantified by PCR and deep sequencing of the gRNA pairs before and after selection.
Adopting the framework of genome-wide CRISPR/Cas9 screens, we synthesized, cloned, and lentivirally delivered thousands of programmed gRNA pairs to cells at a low MOI. Each gRNA pair targets nearby sites, effectively leveraging CRISPR/Cas9’s ability to generate kilobase-scale deletions when NHEJ-mediated repair of two double-stranded breaks results in excision of the intervening DNA segment. In total, we designed and introduced gRNA pairs programming 4,342 overlapping ∼1 and ∼2 kb deletions that tiled a 206 kb region centered on HPRT1. We used 6TG to select for cells that had lost HPRT1 function. By quantifying gRNA pairs both before and after 6TG selection and then directly genotyping putatively important deletions by long-read sequencing, we were able to identify programmed deletions that significantly compromised HPRT1 expression and function.
Material and Methods
Tissue Culture
HAP1 cells were purchased from Horizon Discovery and cultured in Iscove’s modified Dulbecco’s medium with L-glutamine and 25 mM HEPES (GIBCO). The HAP1 cell line was derived from the near-haploid KBM7 line (male cells of chronic myelogenous leukemia origin) by the introduction of induced pluripotent stem cell factors. Despite the cell line’s male origin, HAP1 cells no longer hold a Y chromosome.36 HEK293T cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium with high glucose and sodium pyruvate (LifeTechnologies). Both media were supplemented with 10% fetal bovine serum (Rocky Mountain Biologicals) and 1% penicillin-streptomycin (GIBCO) and grown with 5% CO2 at 37°C.
gRNA Library Design
To generate a list of gRNAs, we identified all 20 bp protospacers followed by a 5′-NGG PAM sequence from chrX: 133,507,694–133,713,798 (UCSC Genome Browser build hg19). We then excluded protospacers that had a perfect sequence match elsewhere in the genome and scored the remaining gRNAs for both on-target and off-target activity. We considered off-target sequences that had five or fewer mismatches to the putative gRNA and calculated an aggregate off-target score by using the method of Hsu et al.37 In addition, we scored each site for on-target efficiency.38 We matched final deletion pairs by using spacers that did not contain BsmBI restriction sites, were not predicted to have off-target hits in other 6TG resistance genes or in KBM7 essential genes (the HAP1 parental cell line), were greater than 25 bp apart, were further than 50 bp from an exon, and passed on-target (above 10) and off-target (above 25) thresholds. Contrastingly, the library of individual gRNAs included all of the spacers targeting the same region, excluding those predicted to have 2,000 or more off-targets or to have off-targets with four or fewer mismatches within the targeted HPRT1 region.
Building the Library of gRNA Pairs
This library cloning method was developed in parallel with similar recently published methods39 and was modified from the GeCKO individual-gRNA cloning scheme.35, 40 First, the lentiGuide-Puro backbone (Addgene 52963) was digested with BsmBI (FastDigest Esp3I, Thermo Fisher Scientific) and gel purified. The paired spacers (flanked with lentiGuide-Puro overlap sequences) were synthesized twice on a microarray (CustomArray) such that each pairing was represented in both possible orders (Figure S1).
To ensure quality of array synthesis, we amplified 1 ng of the oligonucleotide (oligo) pool with Kapa HiFi Hotstart ReadyMix (KHF, Kapa Biosystems) and ran it on a gel to confirm that the oligos were of the expected 108 bp length. After PCR purification with Agencourt AMPure XP beads (Beckman Coulter), the amplicon was cloned into lentiGuide-Puro with In-Fusion HD Cloning Plus (Clontech) and transformed into Stable Competent E. coli (NEB C3040H) for minimizing repeat-based recombination of the lentivirus. This ensuing library (lentiGuide-Puro-2×Spacers) now contained each pair of spacers but was still missing the additional gRNA backbone and PolIII promoter.
We next cloned in the additional gRNA backbone and H1 promoter between each spacer pairing to enable expression of the two independent gRNAs. We ordered the gRNA-backbone-H1-promoter fragment as a gBlock (IDT) with flanking BsmBI sites to allow ligation into the BsmBI-digested lentiGuide-Puro-2×Spacers library. The gBlock and the lentiGuide-Puro-2×Spacers were each digested with BsmBI, purified, ligated together with Quick Ligase (NEB M2200S), and transformed into Stable Competent E. coli for the generation of a final lentiGuide-Puro-2×gRNA library.
To prevent bottlenecking of the library, we performed these cloning steps with enough replicates at high efficiency to maintain a minimum of 20× average library coverage (in relation to the expected library complexity). Sequencing of the lentiGuide-Puro-2×gRNA library revealed 97.8% retention of diversity from the designed paired spacers. However, 16% of library reads held unprogrammed, interswapped pairs. 88.5% of these swaps were seen only in a single read, implying that a more likely cause was template switching during either PCR or cluster generation. For all experimental analyses, only reads of gRNA pairs that perfectly matched programmed pairs were considered.
Building the Library of Individual gRNAs
The spacers of this library were similarly synthesized on an array, amplified, and purified as above. The lentiGuide-Puro backbone was linearized as above, and the library was cloned into it with NEBuilder HiFi DNA Assembly Master Mix (NEB). This plasmid was transformed into Stable Competent E. coli, generating enough transformants for 30× average coverage. This method produced 98.5% retention of complexity from the designed array.
Lentiviral Library Production, Delivery, and 6-Thioguanine Selection
We produced lentivirus with Lipofectamine 3000 (Life Technologies) to transfect HEK293T cells with the lentiviral vector libraries made above and third-generation packaging plasmids (pMDLg/pRRE Addgene 12251, pRSV-Rev Addgene 12253, and pMD2.G Addgene 12259). Supernatant was collected 72 hr after transfection, centrifuged at 300 rcf for 5 min for the removal of cell debris, and passed through a 0.45 μm syringe filter.
For the creation of a monoclonal HAP1 cell line stably expressing Cas9, HAP1 cells were transduced with lentivirus produced with lentiCas9-Blast (Addgene 52962), selected with 5 μg/mL Blasticidin (Thermo Fisher Scientific), and single-cell sorted via fluorescence-activated cell sorting (FACS).
HAP1-Cas9-Blast monoclonal cells were plated to be at 30% confluency on the day of lentiviral gRNA-pair transduction. For transduction, 5% of the recipient cells’ media was replaced with filtered virus, limiting the MOI to <0.3. Media were changed after 24 hr, and selection for transduced cells began 48 hr after transduction. Puromycin was added at 2 μg/mL for 2 days for the assessment of the percentage of cells transduced, and then cells were maintained in 1 μg/mL for 5 more days.
After puromycin treatment, an initial population of cells was collected. Selection for loss of HPRT function was performed by application of 5 uM 6TG to the remaining cells at <50% confluency for 7 days. An additional concern was that minor gene-expression changes caused by ScanDel-mediated mutations in regulatory elements might not be strong enough to confer resistance. To mitigate this, we used the lowest dosage of 6TG that completed HAP1 selection after 7 days. 6TG concentrations of 6–60 μM are reported in the literature to achieve effective selection in this time frame, depending on cell type.41, 42 We tested our monoclonal HAP1-lenti-Cas9-Blast line at concentrations just below this range (1, 2.5, and 5 μM 6TG). After 7 days, the 5 μM treatment had no readily identifiable surviving cells, whereas the 2.5 μM treatment retained a sparse population, and the 1 μM treatment produced appreciably more outgrowing colonies. On the basis of these results, we proceeded with selections by using 6TG at 5 μM for 7 days. Enough cells were transduced and sampled at each time point to maintain a minimum 2,000× average coverage of the library in each population.
Sequencing of the baseline (i.e., pre-6TG) population revealed that 98.4% diversity of the lentiGuide-Puro-2×gRNA library was preserved from replicate 1, and replicate 2 retained 78.8%. Because our deletions highly overlapped, we proceeded with replicate 2 because all base pairs were interrogated despite the lower diversity. We observed 95.6% retention of programmed library diversity in replicate 1 of the individual-gRNA plasmid library and 71.2% of replicate 2.
Interswapped gRNA pairs were observed in 35.5% of reads from the baseline pre-6TG sample. This is an increase from the 16% observed in reads from the lentiGuide-Puro-2×gRNA plasmid library. This suggests additional template switching during the library’s amplification from gDNA, which would require more cycles of PCR. However, because we directly sequenced each gRNA spacer as a readout instead of using barcoded libraries30 and only took exact sequence matches, this did not pose a problem.
gRNA Library Amplification and Sequencing from HAP1 Cells
gDNA was extracted from the cells sampled before and after 6TG selection with the DNeasy Blood & Tissue kit (QIAGEN). KHF was used for all amplification steps. The libraries were initially amplified from a minimum of 6 μg of gDNA divided across thirty 50 μL reactions, ensuring sampling of about two million haploid genome equivalents at each time point. We performed two additional PCRs, and AMPure bead purification between each reaction, to add sequencing adapters and sample indices to the amplicon. We optimized amplification conditions by qPCR to minimize overamplification of the construct.
Sequencing was performed on an Illumina MiSeq with a 50-cycle kit. Read 1 and the Illumina index read were used for sequencing the two gRNAs in the paired-gRNA construct before paired-end turnaround, and read 2 was used for sequencing the 9 bp sample index.
Calculation of a Selection Score Assignment per Base Pair
Custom Python scripts counted tallies of gRNAs (for experiments with the library of individual gRNAs) or gRNA pairs before and after selection. These counts were normalized to the total number of reads per sample. We calculated an enrichment ratio for each gRNA pair by dividing its normalized read count after selection by its read count before selection. A selection score is the log10 of the enrichment ratio: log10(after/before). If a gRNA or gRNA pair was absent before selection, it was excluded from further analysis. Any gRNA pairs that used a self-paired gRNA with an independent selection ratio > 0 were also excluded from further analysis.
If a gRNA pair is absent after 6TG selection, its calculated selection score will be a negative number that is relatively large in magnitude and somewhat arbitrarily determined by the number of pre-selection reads. Thus, to limit the contribution of these scores to average measurements derived from many independent deletions, we set a minimum selection score equal to the middle of the bimodal distribution between the positively and negatively selected deletions of each replicate (Figure S2). For example, in ScanDel replicate 1, if the log10 value of a selection score was less than −0.35, that gRNA pair’s score was set to −0.35. We assigned each individual base pair a per-base-pair selection score by taking the mean of all deletions programmed to cover that base pair. The per-base-pair score was normalized to the median score for all positive scores in that replicate. We averaged the per base-pair selection score of each replicate to get the final selection score per base pair. Per-base-pair scores were uploaded as a bedgraph for visualization on the UCSC Genome Browser.
For the individual-gRNA mutagenesis screen, we calculated selection scores per base pair similarly by assuming that a 10 bp deletion was made by each gRNA queried. If a base pair was scored at the minimum negative threshold in one screen, it was given that value for the consensus selection score of the two replicates.
Bulk ATAC-Seq of HAP1 Cells
Two biological replicates were separately maintained (on 10 cm dishes and split 1:10 three times per week) and processed separately. Chromatin accessibility in the HAP1 cell line was profiled with the ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing) protocol43 with slight modifications. The media for 10 cm plates of confluent HAP1 cells were aspirated and replaced with 2 mL of ice-cold lysis buffer (CLB+; made as described in Buenrostro et al.43 but supplemented with protease inhibitors [Sigma cat. no. P8340]). Cells were incubated on ice for 10 min in CLB+ and then were dislodged with a cell scraper and transferred to a 15 mL conical tube and pelleted at 500 rcf for 5 min at 4°C. Nuclei were re-suspended in 1 mL of CLB+ and counted on a hemocytometer. 50,000 nuclei in 22.5 μL of CLB+ were combined with 2.5 μL of TDE1 enzyme and 25 μL of TD buffer (Illumina). Tagmentation conditions were as described in Buenrostro et al.43 (37°C for 30 min). After MinElute purification into 10 μL EB buffer (QIAGEN), 5 μL of tagmented DNA was amplified in 25 μL reactions for 12 cycles with NEBNext Master Mix (NEB). Reactions were monitored with SYBR Green for ensuring that samples were not overamplified. PCR products were cleaned once with a QiaQuick PCR Cleanup Kit (QIAGEN) and once with 1× AMPure beads (Agencourt). The quality of the library was assessed on a 6% TBE gel, and the yield was measured by a Qubit (1.0) fluorometer (Invitrogen).
Samples were sequenced on two paired-end Illumina NextSeq 500 runs. Read lengths were 2 × 75 bp for the first run and 2 × 151 bp for the second run, so the second run was truncated to 75 bp. Sequencing reads were also trimmed for readthrough of adaptor sequences and quality with Trimmomatic44 (“NexteraPE-PE.fa:2:30:10:1:true TRAILING:3 SLIDINGWINDOW:4:10 MINLEN:20” parameters) and then mapped to the 1000 Genomes integrated reference genome “hs37d5” with bowtie245 and the “-X 2000 -3 1” parameters. Only properly paired and uniquely mapped reads with a mapping quality above 10 were retained (“samtools -f3 -F12 -q10”). Reads mapping to the mitochondrial genome and non-chromosomal contigs were also filtered out. In addition, duplicate reads were removed with Picard. After checking quality-control metrics on the individual replicates, we combined reads from the two libraries for downstream analysis. Hypersensitive sites were called (at a 1% false-discovery rate) with the Hotspot algorithm.46
Validation and Direct Genotyping of Positive Signal from the Screens
gRNA pairs that drove the ScanDel signal surrounding HPRT1’s first exon were cloned into simple lentiGuide-Puro-2×gRNA libraries. The transcription start site (TSS) ScanDel validation library contained four pairs, and the intron 1 library contained five (Tables S1 and S2). For the TSS sub-library validating the individual-gRNA screen, ten gRNAs were cloned into lentiGuide-Puro (Table S3). These constructs were lentivirally delivered to HAP1-Cas9-Blast cells and selected with 6TG, and gDNA was extracted as described above.
Because the expected deletions could remove up to 3 kb, the loci were sequenced with a Pacific Biosciences RSII (University of Washington PacBio Sequencing Services, P6C4 chemistry, RSII platform). To prepare libraries for PacBio sequencing, we amplified the TSS- or intron 1-targeted regions from 800 ng of gDNA each by using four 50 μL KHF reactions with primers adding sample indices and SbfI or NotI cut sites. The purified amplicons (Zymo Research DNA Clean & Concentrator-5) were digested with SbfI-HF (NEB) and NotI-HF (NEB), leaving sticky ends. 5′-phosphorylated SMRT-bell hairpin oligos (IDT) containing the PacBio priming site, hairpin-forming sequence, and resulting sticky ends for either SbfI or NotI were annealed by heating to 85°C and snap frozen in 10 mM Tris 8.5, 0.1 mM EDTA, and 100 mM NaCl. These were ligated at 10× molar excess to the digested amplicons, destroying the restriction site once attached. For the removal of undigested amplicons and primers, this ligation was performed in the presence of further SbfI and NotI and was followed by treatment with Exo7 (Affymetrix) and Exo3 (Enzymatics).
Only reads with over five circular consensus sequence passes and containing the expected first twelve 5′ and 3′ base pairs of the amplicon were used for further analysis. Reads positive for complex inversions (≥100 bp) were removed from the library by the Waterman-Eggert algorithm with match, mismatch, gap open, and gap extend scores of 2, 10, 10, and 5, respectively.47 The resulting reads were then aligned to the amplicon reference with the NEEDLEALL48 aligner with a gap-open penalty of 10 and a gap-extension penalty of 0.5. Insertions were required to start within a window of 5 bp up- or downstream of the putative cut site. Deletions were required to either start or end within the same 10 bp window or span the window. Reads that carried the same edit pattern were collapsed into haplotypes, and figures were generated with a custom D3 script.
Comparing Deletion Rates of U6-H1 and U6-U6
We chose two protospacers to program a 365 bp deletion within the second intron of HPRT1 and cloned their spacers into a U6-H1 construct and a U6-U6 construct (Figure S3). Virus was produced and delivered to cells, which were selected with puromycin, and gDNA was extracted as described above. The locus was amplified in four successive rounds of nested PCR. The first reaction was only three cycles and included a forward primer with a 10 bp unique molecular index (UMI). The second reaction amplified any UMI-tagged fragments. The third and fourth reactions added sample indices and Illumina flow-cell adapters. The products were cleaned by AMPure between each reaction at a concentration that would lose primer dimer but retain the smaller deletion-holding fragments and were sequenced on a MiSeq. Any reads that contained the same UMI or edit pattern were collapsed by custom scripts, and their alignments were visualized with the same D3 script as above.
Results
Development of ScanDel
In genome-wide CRISPR/Cas9 screens, a gRNA library is lentivirally delivered to a large pool of cells at a low MOI, such that each infected cell is likely to receive only one gRNA.40, 49, 50 With the goal of perturbing the function of the targeted locus, each gRNA induces NHEJ-mediated indels centered at the Cas9-mediated cleavage position within the target sequence. However, given the small and variable length of indels, the robustness of perturbation is inherently limited, particularly when non-coding sequences in which frameshifts are irrelevant are being targeted. To instead program a kilobase-scale deletion in each cell, we devised the following approach (Figure 1). First, gRNA pairs are designed to program specific deletions (each gRNA specifies one of the deletion’s boundaries; Figure 1A), and the corresponding pairs of 20 bp spacers are synthesized in cis on a microarray (Figure 1B). Second, the paired spacers are inserted into the lentiGuide-Puro plasmid between the U6 promoter and the gRNA backbone. Third, a second gRNA backbone and a second RNA polymerase (Pol) III promoter (H1 or U6) are inserted between the paired spacers. Fourth, libraries of gRNA pairs are lentivirally delivered to a large pool of cells at a low MOI, such that each cell receives a pair of gRNAs that program a single deletion (Figure 1C). Finally, analogous to conventional genome-wide CRISPR/Cas9 screens, deep sequencing of the integrated gRNA pairs is used as a surrogate measure of the prevalence of each programmed deletion in a population of cells (e.g., before and after the cells have been subjected to functional selection), thus capturing the phenotypic consequences of individual deletions.
As an initial test of our paired guide system, we compared the efficacy of using two different promoters for the two guides (a U6-H1 system) with that of using two copies of the same promoter (U6-U6). We tested these lentiviral gRNA-pair expression constructs by targeting the same genomic site for deletion with each system (Figure S3). We performed PCR amplification of the site with UMIs in order to minimize biases related to amplicon size (see Material and Methods). The U6-H1 system induced more programmed deletions than the U6-U6 system (20% versus 10% of reads from cells 1 week after transduction). The U6-H1 system has several advantages (e.g., it avoids recombination between the two U6 promoters during cloning and has a unique primer design for deep sequencing of each gRNA), and we therefore proceeded with it.
An important caveat for ScanDel, in relation to conventional gRNA cell-based screens, is that deletions programmed by gRNA pairs occur only in a minority of cells;51, 52 the other major outcomes are small NHEJ-mediated indels at one or both gRNA-targeted sites. For example, in our test of the U6-H1 system, the programmed deletion was found in 32% of cells that had any edit, whereas the remaining edited cells were mutated at one or both gRNA-targeted sites but retained the intervening sequence. Although this complicates interpretation, the problem can be overcome by a robust functional assay in conjunction with multiple, independent gRNA pairs that query the same genomic region, as well as by the inclusion of unpaired gRNA controls to ensure that observed effects do not occur with the individual gRNAs that make up each pair (but rather are dependent on the presence of both gRNAs).
Application of ScanDel to Survey the 206 kb Region Surrounding HPRT1
With the goal of investigating the potential of non-coding mutations to compromise its function, we applied ScanDel to a 206 kb X chromosome region centered on HPRT1 (Figures 1A and 2A). We designed pairs of gRNAs that programmed deletions tiling across the 206 kb region, including tiles that overlapped HPRT1 exons, in order to allow coding regions to serve as positive controls. Given that deletion length has been shown to affect deletion rate,51 deletions were programmed to be consistently either ∼1 or ∼2 kb in length (Figure 1A). This design resulted in 4,342 programmed deletions that tiled across the region, and they collectively covered each base pair a median of 27 times (Figure 2B). Testing each base pair with numerous independently programmed tiling deletions is expected to reduce noise and also increase resolution (given that all successfully made deletions tiling a critical regulatory element should exhibit positive selection). However, to guard against the possibility that the effects of individual gRNAs could confound analysis (e.g., via off-target mutations or on-target small ∼10 bp indels), we also included all spacers in the library as pairs with themselves (“self-pairs”; Figure 1B, inset; Figure S1). Additionally, we included 330 negative-control gRNA pairs not expected to survive 6TG selection, because they program deletions in non-genic regions far from HPRT1 or use spacers made of random sequence not present in the reference genome (hg19).
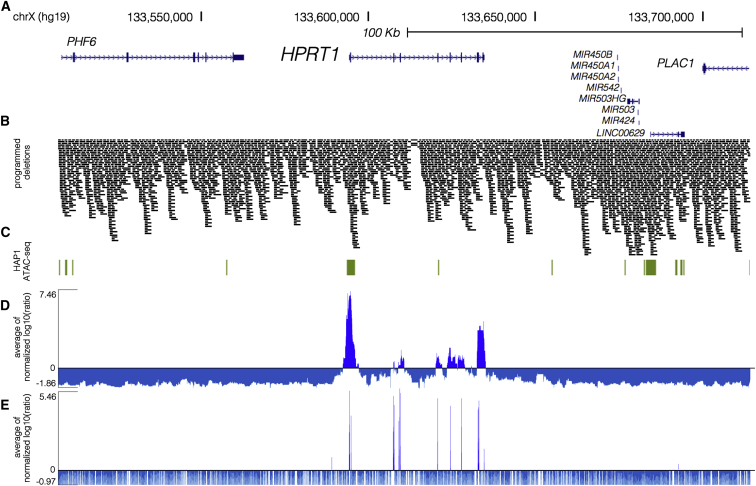
Figure 2.
High-Coverage ScanDel Library across the HPRT1 Locus Reveals a Paucity of Critical Distal Regulatory Elements
(A) Deletions were programmed across 206.1 kb of the HPRT1 locus and its surrounding sequence (chrX: 133,507,694–133,713,798, hg19; UCSC Genes track in blue).
(B) A total of 4,342 overlapping 1 or 2 kb deletions were programmed (see Figure 1A) to tile across the locus such that each base pair was interrogated by a median of 27 independently programmed deletions. A high density of repeat elements resulted in reduced coverage of a region within intron 3 of HPRT1. Deletions are represented by black bars spanning the gRNA pair’s programmed cut sites.
(C) HAP1 ATAC-seq hotspots (green) indicate regions of open chromatin in the cell line. Of note, a hotspot extends 600 bp upstream and 1.6 kb downstream of exon 1.
(D) ScanDel scores were assigned to each base pair as the average of all selection scores (log10(after/before)) for gRNA pairs that programmed deletions to span that base pair (Material and Methods). If a gRNA pair used a spacer that was positively selected on its own as a self-pair, that gRNA pair was removed from the analysis. Given that depleted gRNAs are usually completely absent after 6TG, their negative scores are of arbitrary negative magnitude. To avoid over-weighting negative values, we determined a minimum score from each replicate’s gRNA-pair score distribution (Figure S2), and scores below it were set at this minimum. For each biological replicate, the base pair’s score was normalized to the replicate’s median of positive scores. The average of the two biological replicates’ normalized scores for that base pair is displayed (positive scores in royal blue and negative scores in blue-gray).
(E) An individual-gRNA mutagenesis screen of the same region was also performed and covered only ∼70% of bases in the region as a result of the sparsity of high-quality designable spacers. Individual base pairs were scored on the basis of nearby cut sites under the assumption that each gRNA queries a ∼10 bp region. The plotted scores were calculated as in (D) (positive scores in royal blue and negative scores in blue-gray).
The gRNA-pair library was array synthesized, cloned, and delivered via lentiviral infection to HAP1 cells in replicate (Figures 1B and 1C). Cell populations were sampled before and after 1 week of the 5 μM 6TG selection, and PCR amplification and deep sequencing of gRNA pairs were used to quantify abundance at each time point. The functional selection score was calculated as the log10 ratio of normalized read counts after selection to those before 6TG treatment (“selection score” as log10(after/before 6TG)). Positively scoring self-paired spacers were flagged, and gRNA pairs that used these flagged spacers were excluded from further analysis (11% of pairs in replicate 1 and 3% of pairs in replicate 2). To integrate signal from overlapping programmed deletions, we calculated a “per-base-pair” metric as the mean of selection scores of all deletions overlapping a given base (Figure 2D and Material and Methods). This per-base-pair score across the HPRT1 locus was well correlated between biological replicates (Pearson: 0.708; Figure S4). Importantly, none of the negative-control gRNA pairs that were sampled in each of the two replicates were positively selected in both experiments (Figure S5).
Crucially, all nine HPRT1 exons exhibited strong functional scores, confirming the sensitivity of ScanDel as applied here to detect sequences essential to HPRT1 function (Figure S6). However, all reproducibly positive non-coding signal across the 206 kb region was immediately proximal to an HPRT1 exon. This result suggests that no distal regulatory element in the 206 kb region is essential to HPRT1 expression in HAP1 cells.
Near exons, non-coding regions exhibiting positive signal did so even when deletions that also overlapped the exons themselves were excluded from the analysis (Figure S6D). This suggested the presence of essential, proximal regulatory sequences. We noted that the positively scoring regions immediately upstream and downstream of the first exon overlapped a region of open chromatin identified by ATAC-seq in HAP1 cells, supporting the region’s role in gene regulation (Figure 2C and Figures S6 and S7). Together, these observations motivated us to attempt validation experiments for this region with the goal of directly confirming which deletions of putative regulatory elements were impairing HPRT1 function (Figures 3A and 3E).
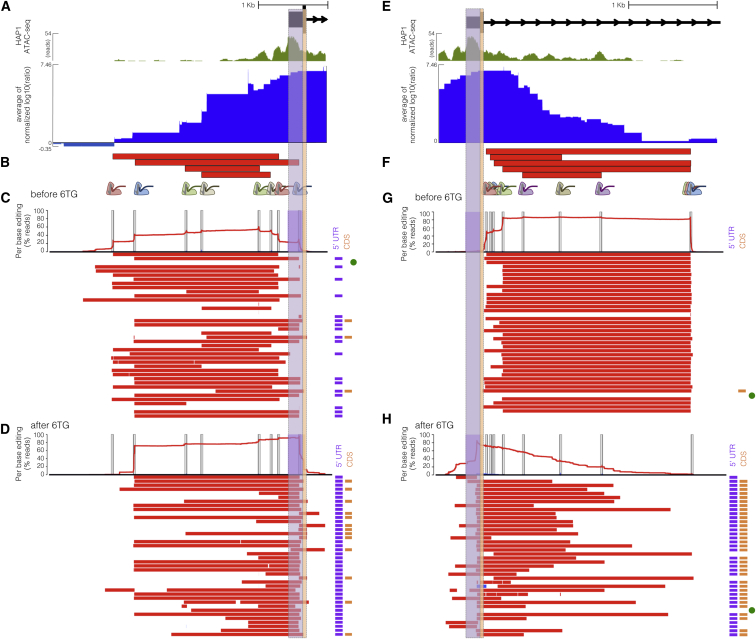
Figure 3.
Long-Read Sequencing of Edits Derived from Exon-Proximal ScanDel gRNA Pairs Reveals Rare, Unprogrammed, Exon-Interrupting Deletions That Drive Selective Effects
(A) A putative promoter is implicated by open chromatin (HAP1 ATAC-seq broad peaks, green) surrounding exon 1 of HPRT1 (UCSC Genes, black). ScanDel signal in the 2 kb upstream of HPRT1 also suggests the possibility of critical regulatory sequences in this region (chrX: 133,591,603–133,594,626, hg19; blue as in Figure 2D). The 5′ UTR and coding regions of exon 1 are highlighted in purple and orange, respectively.
(B) Four gRNA pairs targeting the promoter were cloned as a small pool, delivered, and selected with 6TG to enable sequencing of the edited locus (programmed deletions are displayed as red bars). A 3 kb region was amplified and sequenced with long reads (Pacific Biosciences).
(C) The chart at the top displays the per-base percentages for deletions (red) and insertions (blue), and target sites are indicated by vertical gray bars. Horizontal bars show the edits found on each haplotype (red, deletions; blue, insertions; ranked by decreasing prevalence). All programmed deletions, in addition to rare, unexpected deletions, were abundant before 6TG treatment. The notations to the right indicate whether the edits interrupt the TSS or 5′ UTR (purple bar) and/or coding sequence (orange bar). The unedited haplotype is marked with a green dot. Of note, PCR and sequencing on the PacBio RSII were biased toward smaller fragments, limiting accurate quantitative comparison of read counts from differently sized edits.
(D) Haplotypes from 6TG-selected cells are plotted as in (C), revealing that only edits interrupting the TSS or 5′ UTR survive selection and that no programmed or “promoter only” deletions survive selection.
(E) Open chromatin (green as in A) and ScanDel signal suggest the presence of critical non-coding regulatory sequences in the first ∼2.7 kb of intron 1 (chrX: 133,593,871–133,596,998, hg19).
(F) Five gRNA pairs that drove the signal in this intronic region were cloned and selected with 6TG as a small pool, as in (C).
(G) A 3.1 kb region spanning the most-5′ part of intron 1 was amplified and sequenced from cells sampled before 6TG selection. Haplotypes and per-base editing rates are diagrammed as in (C).
(H) Post-6TG selection haplotypes from the intron-1-targeted cells are plotted as in (G), revealing that the vast majority of surviving edits disrupt the exon. Two edited haplotypes do not interfere with the exon, but these are present at approximately the level of unedited haplotypes, suggesting that 6TG resistance in these cells is caused by mutations elsewhere.
Direct Genotyping of Deletions That Survive Functional Selection
With the goal of validating the positive signal upstream of the first exon, we repeated the experiment with a small pool of four gRNA pairs targeting the putative HPRT1 promoter (Figure 3B). We then amplified 3 kb of this region by PCR and performed long-read sequencing of the amplicons (Pacific Biosciences). As expected, before 6TG selection, the programmed deletions were all well represented in the population, although deletions with boundaries deviating from Cas9 cut sites (i.e., “unprogrammed”) were also detected (Figure 3C). However, after selection with 6TG, deletions with unprogrammed boundaries predominated, including those unseen before 6TG and those that extended beyond the TSS (Figure 3D). The fact that these initially rare deletions were strongly selected (whereas 2 kb promoter deletions that did not cross the TSS were not) suggests that even relatively proximal sequences upstream of the HPRT1 TSS are not strictly essential for expression. On the basis of the results of these validation experiments, we conclude that only a narrow window of non-coding sequence immediately upstream of the TSS and 5′ UTR is required for HPRT1 expression.
We next sought to validate the positive signal downstream of the first exon. To do so, we again repeated the experiment with a small pool of just five gRNA pairs targeting the first ∼2.7 kb of intron 1 (Figure 3F). We then amplified the region and again performed long-read sequencing of the amplicons (Pacific Biosciences). As with the promoter, the programmed deletions were all well represented before 6TG selection, although deletions with unprogrammed boundaries were also detected at a low rate (Figure 3G). After selection, deletions with unprogrammed boundaries predominated again, particularly those that extended into the first exon, thereby disrupting coding sequences (Figure 3H). A low rate of non-exonic deletions survived after 6TG, but these were present at the same level as unedited reads, implying that there could be some other explanation for 6TG resistance in these cells. Thus, as with the promoter, the positive signals that we originally observed for deletions in the first intron were most likely a result of the positive selection of rare “on-target-but-with-incorrect-boundaries” deletions that extended into the first HPRT1 exon.
An Individual-gRNA Screen of the Same Region for Comparison with ScanDel
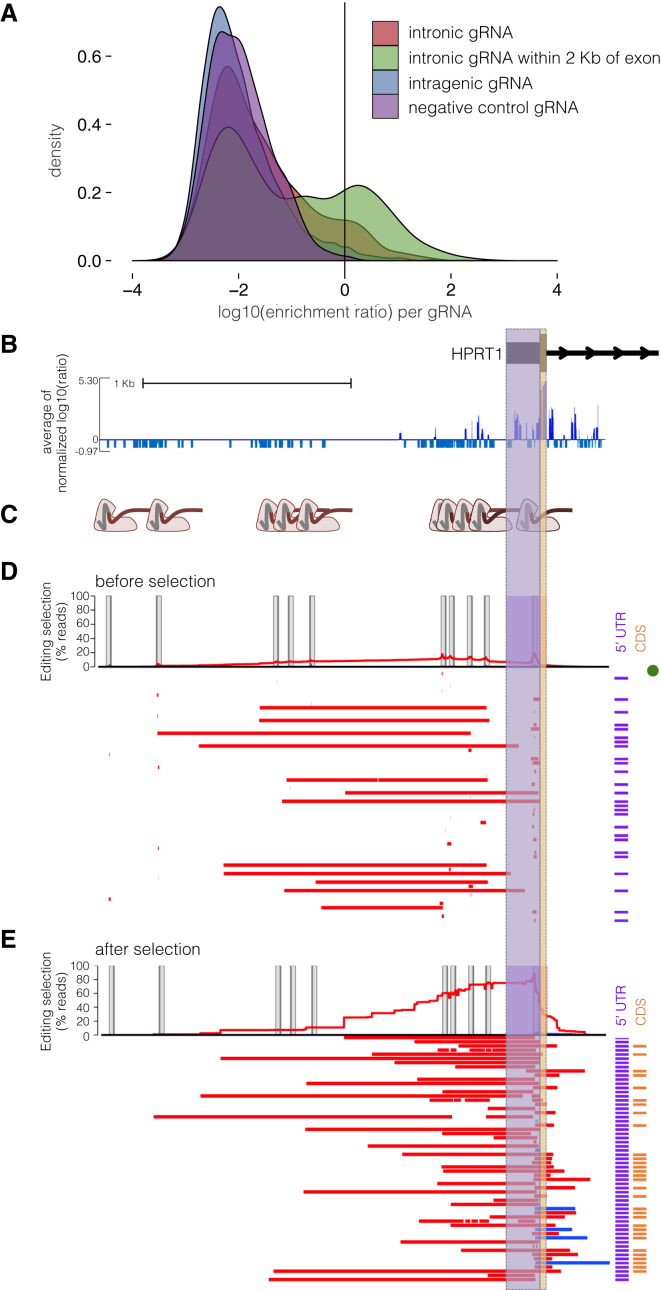
We next compared our ScanDel results against a more conventional screen relying on only individual gRNAs16, 17, 18, 19, 21 (Figure 2E). For this, we cloned a second lentiviral library consisting of 12,151 individual gRNAs targeting the same 206 kb region and assayed HPRT function in HAP1 cells as previously. Under the assumption that each individual gRNA potentially disrupts a ∼10 bp region, this experiment at best interrogates ∼70% of bases within the 206 kb region as a result of the sparsity of PAM sites (by comparison, ScanDel covers each base pair in the entire locus at a median ∼27-fold redundancy). 86% of exon-targeting gRNAs were positively selected, and exonic selection scores were well correlated between biological replicates (Pearson: 0.781). Of 612 negative-control gRNAs, none that were sampled in each replicate were positively selected in both experiments (Figure S8). In non-coding sequence, scores were poorly correlated between biological replicates, and there was a paucity of reproducible, positively selected signal (Pearson: 0.156; Figure S9).
Notably, we did observe a greater proportion of positively scoring gRNAs in the vicinity of exons—i.e., whereas only 2% of intergenic gRNAs were positively selected, 7.5% of deep intronic (>2 kb away from an exon boundary) and 20.5% of proximal intronic (<2 kb from an exon boundary) gRNAs were positively selected (Figure 4A). Given our earlier observation with ScanDel of rare “on-target-but-with-incorrect-boundaries” deletions that were confounding when targeting near exon boundaries, we next performed similar validation experiments on individual gRNAs that targeted non-coding sequences near exons (Figure 4B). We chose ten gRNAs in the HPRT1 promoter region (Figure 4C) and repeated the individual-gRNA experiment with a small pool of just these ten gRNAs, again by using long reads (Pacific Biosciences) to sequence the locus before (Figure 4D) and after (Figure 4E) 6TG selection. Similar to our results with ScanDel in this region, the only mutations that survived 6TG selection were initially rare deletions whose boundaries extended past the TSS and into the 5′ UTR and/or coding sequence (Figure 4D). This result strongly underscores that caution should be exercised in the interpretation of results from CRISPR-based screens of non-coding regions, whether performed with individual gRNAs or gRNA pairs, and the importance of sequencing-based validation of edited regions in the context of such screens.
Figure 4.
Direct Genotyping of Edits from an Individual-gRNA Mutagenesis Screen Also Reveals Rare, Unexpected Edits Disrupting Exon 1 of HPRT1
(A) A greater proportion of gRNAs targeting non-coding sequence within 2 kb of exons were positively selected in an individual-gRNA screen across the HPRT1 locus (data shown from replicate 1; Figure 2E). Each gRNA was assigned a score equal to the log10(after/before 6TG).
(B) gRNAs that target upstream of the transcriptional start site were positively selected. The 2.4 kb region sequenced for genotype validation (chrX: 133,592,240–133,594,646, hg19), i.e., a zoom-in of data from the whole region in Figure 2E, is also shown.
(C) For validation, ten gRNAs in this 2.4 kb promoter region were cloned into a low-complexity library, delivered to HAP1 cells expressing Cas9, and selected with 6TG. After selection, the 2.4 kb promoter region was amplified for long-read sequencing.
(D) Reads from before 6TG selection are plotted as in Figure 3C. In brief, the per-base percentage of haplotypes that carried a deletion (red) or insertion (blue) is charted. The edits of the most-prevalent haplotypes from long-read sequencing are drawn as colored bars, and the notations to the right indicate whether the edits interrupt the TSS or 5′ UTR (purple) or coding sequence (orange) of exon 1. A green dot signifies the unedited haplotype. Target-site programmed edits are observed and are mainly composed of the expected small indels, in addition to rarely occurring larger deletions. PCR and sequencing on the PacBio RSII were biased toward smaller fragments, limiting accuracy of quantitative comparison of the read-count prevalence of different sized edits.
(E) The most abundant haplotypes from cells after 6TG selection are visualized as in (D). Only mutations that interrupt exon 1 survived 6TG selection.
Discussion
We developed a method that uses CRISPR/Cas9 and pairs of gRNAs to experimentally test the functional consequences of thousands of programmed, kilobase-scale genomic deletions in a single experiment. We applied this method to perform the systematic investigation of the regulatory architecture of a housekeeping gene via editing of the endogenous genome. Upon introducing a set of densely tiling deletions spanning a 206 kb region centered on the gene HPRT1, we found no evidence that any distal regulatory element is critical for its activity, as measured by 6TG sensitivity in HAP1 cells. A screen of this same region with individual gRNAs supported this finding. The dearth of positive selection from disruption of non-coding regions contrasts with the strong positive selection observed from disruption of any exon of HPRT1 either by programmed deletions or individual guides.
HPRT1 is a widely expressed housekeeping gene53 with no eQTLs identified by the Genotype-Tissue Expression Project7 and thus might not require multiple (or any) distal regulatory regions for its expression. The simplest explanation of our results is that sequences immediately proximal to the HPRT1 TSS could be sufficient to confer the level of expression that provides sensitivity to 6TG, such that even if we disrupt distal regulatory elements that subtly modulate expression, they would go undetected by our strong selection. For future applications of ScanDel, implementing more quantitative readouts will be critical. For example, ScanDel is compatible with any functional selection that reliably separates cells on the basis of gene expression (e.g., by knocking in GFP to a locus of interest and then using FACS to stratify ScanDel-edited cells on the basis of expression). Such quantitative readouts could facilitate validation of the many candidate regulatory elements (and cognate target-gene assignments) nominated by eQTL and functional genomics studies.54, 55 We anticipate that the application of ScanDel to non-housekeeping genes in conjunction with a more quantitative readout is likely to identify more regulatory elements than found for HPRT1, especially for genes that play key roles in development and cell-fate determination.
Another possibility, albeit an unlikely one, is that critical regulatory elements for HPRT1 lie outside of the 206 kb window that we surveyed. For example, the gene resides at the terminus of a ∼300 kb topologically associated locus that spans ∼185 kb beyond our interrogated region in HAP1 cells56 (Figure S10). One could potentially address this by increasing the complexity of the library of programmed deletions in order to densely tile a larger region or by simply increasing the size of each programmed deletion to interrogate more sequence per gRNA pair.
We note that the paucity of regulatory sequences discovered by CRISPR/Cas9-based screening is not exclusive to this study. Collectively, individual gRNA CRISPR/Cas9 screens have surveyed over a megabase of prioritized non-coding sequences, but only a handful of gRNAs tested have robust phenotypic effects that validate.16, 18, 19, 20, 21 One explanation is that the assays being used are insufficiently sensitive and fail to detect modest regulatory effects. This could be addressed through the implementation of more quantitative assays.
A second explanation is that as implemented, genome editing has poor sensitivity as a result of redundancy in mammalian gene regulation. Redundancy of transcription factor binding sites within enhancers could prevent ∼1–10 bp indels introduced by individual gRNAs from sufficiently disrupting function. Indeed, this was part of the motivation for developing ScanDel, whose programmable kilobase-scale deletions exceed the size of enhancers. Although we did not identify distal enhancers, the essentiality of the TSS and portions of the 5′ UTR in our assay was detected primarily by deletions substantially larger than 1–10 bp (Figures 4D and 4E), suggesting that libraries of gRNA pairs will be effective for enhancing sensitivity. However, there could also be redundancy among sets of distal regulatory elements, a question that can be fully addressed only by combinatorial perturbations.
A third explanation is that gene expression levels depend in part on historical events, such that disruption of an enhancer in a differentiated cell line would not result in the same outcome as disrupting the same enhancer before differentiation. This could be potentially addressed through lentivirally mediated genome-editing steps in stem cells and subsequent differentiation to a cell type of interest. Any differences in functional consequences that depend on the timing of mutation would be of great interest.
Our results also provide a cautionary example of the importance of validation by direct genotyping in the context of CRISPR/Cas9-based screens of non-coding sequences. NHEJ generates a wide assortment of mutations, and strong selections could recover rare editing outcomes. For example, whereas targeting regions adjacent to exons might have been interpreted to reflect the presence of critical proximal regulatory elements, validation experiments using a long-read sequencer showed that this signal was caused by rare deletions that extended into exonic sequence. Detecting many of these unexpected events would have been difficult had we been relying solely on a short-read sequencing platform to genotype editing outcomes. Additionally, validating CRISPR/Cas9-based screens by assessing selection for specific edited haplotypes adds biological information. Here, with long-read genotyping, we were able to identify a set of variable deletions that either did or did not drive selection, thus enabling greater resolution (Figures 3C and 3D).
We also note that in experiments relying on pairs (or more) of gRNAs to program deletions, it is critical to include controls that quantify the effects of the individual gRNAs making up these pairs, because these can have direct or off-target effects that might be misinterpreted as being consequent to the programmed deletion. While this manuscript was in preparation, Zhu et al. published a study that similarly used gRNA pairs to program deletion of a large number of lncRNAs, as well as subsequent phenotyping for cellular growth.30 Although the results are of great interest, these important controls were not included for the vast majority of spacers used. It will also be important to confirm the validity of each of this screen’s findings through direct genotyping.
Even with the aforementioned open questions and remaining technical hurdles, it is critical that we continue to advance and apply methods for multiplex perturbation of the regulatory landscape with genome editing. The importance of experimental perturbation is highlighted by our results. The non-coding region surrounding the first exon of HPRT1 resides in open chromatin in this cell line (Figure 2 and Figure S6), yet our results with ScanDel and subsequent validation experiments indicate that the essential regulatory region is only a small part of the broader ATAC-seq peak. Perturbing the endogenous genome represents a highly complementary approach to the more classic strategy of reporter assays,57, 58 in which short sequences are tested for their regulatory potential on an episomal vector. Of note, the results of early reporter-assay-based tests of potential regulatory sequences flanking HPRT1 are largely consistent with our findings but also identify three sequences that are immediately proximal to the first or second exon and are critical for episomal HPRT1 expression.28, 29 Although this discrepancy could be due to cell-type or species differences (because two of these elements were required only in mouse embryonic stem cells but not in human cells,28 and the remaining one was tested only in Chinese hamster fibroblasts29), it could also be due to differences in the activity of regulatory elements between episomal assays and genome editing. For example, elements necessary for driving expression of a gene on a plasmid might not be required in the genome, where redundancy is more likely. This underscores the ongoing challenge that genome editing can address: understanding how short sequences with regulatory potential coordinate with one another across endogenous loci to give rise to specific levels of expression.
In summary, ScanDel enables the multiplex characterization of the functional consequences of thousands of programmed, kilobase-scale deletions to the endogenous genome in a single experiment. We applied ScanDel to HPRT1, a housekeeping gene in which disruptive mutations cause Lesch-Nyhan syndrome, by introducing densely tiled 1–2 kb deletions across a 206 kb region encompassing the gene to cover each base pair with median ∼27-fold redundancy. Our results demonstrate that this region lacks distal cis-regulatory elements that are critical for HPRT1 expression. In the future, we anticipate that large-scale perturbation of putative regulatory elements in their endogenous context via methods such as ScanDel will provide further insights into gene regulation and the contribution of non-coding mutations to human disease.
Acknowledgments
For discussion and advice, the authors thank all members of the Shendure Lab, particularly Lea Starita, Andrew Hill, Ron Hause, Seungsoo Kim, Martin Kircher, and Beth Martin. We thank the University of Washington PacBio Sequencing Services core for their assistance. This work was supported by an NIH Director’s Pioneer Award (DP1HG007811 to J.S.), the National Human Genome Research Institute (1R01HG006768 to J.S.), and the National Cancer Institute (1R01CA197139 to J.S. and F30CA213728 to G.M.F.). M.G. is a National Science Foundation Graduate Research Fellow. A.M. was supported by the NIH and National Heart, Lung, and Blood Institute (NHLBI; T32HL007312). D.A.C. was supported in part by the NHLBI (T32HL007828). J.S. is an investigator of the Howard Hughes Medical Institute.
Published: July 13, 2017
Footnotes
Supplement Data include ten figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.06.010.
Contributor Information
Molly Gasperini, Email: gasperim@uw.edu.
Jay Shendure, Email: shendure@uw.edu.
Web Resources
OMIM, http://www.omim.org/
UCSC Genome Brower, http://genome.ucsc.edu
Supplemental Data
References
- 1.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguet F., Brown A.A., Castel S. Local genetic effects on gene expression across 44 human tissues. bioRxiv. 2016 [Google Scholar]
- 8.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coetzee S.G., Rhie S.K., Berman B.P., Coetzee G.A., Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012;40:e139. doi: 10.1093/nar/gks542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M.J., Wang L.Y., Xia Z., Sham P.C., Wang J. GWAS3D: Detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res. 2013;41:W150–W158. doi: 10.1093/nar/gkt456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakabayashi A., Ulirsch J.C., Ludwig L.S., Fiorini C., Yasuda M., Choudhuri A., McDonel P., Zon L.I., Sankaran V.G. Insight into GATA1 transcriptional activity through interrogation of cis elements disrupted in human erythroid disorders. Proc. Natl. Acad. Sci. USA. 2016;113:4434–4439. doi: 10.1073/pnas.1521754113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weedon M.N., Cebola I., Patch A.-M., Flanagan S.E., De Franco E., Caswell R., Rodríguez-Seguí S.A., Shaw-Smith C., Cho C.H., Lango Allen H., International Pancreatic Agenesis Consortium Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claussnitzer M., Dankel S.N., Kim K.-H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diao Y., Li B., Meng Z., Jung I., Lee A.Y., Dixon J., Maliskova L., Guan K.L., Shen Y., Ren B. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korkmaz G., Lopes R., Ugalde A.P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 20.Rajagopal N., Srinivasan S., Kooshesh K., Guo Y., Edwards M.D., Banerjee B., Syed T., Emons B.J., Gifford D.K., Sherwood R.I. High-throughput mapping of regulatory DNA. Nat. Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanjana N.E., Wright J., Zheng K., Shalem O., Fontanillas P., Joung J., Cheng C., Regev A., Zhang F. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A., Findlay G.M., Gagnon J.A., Horwitz M.S., Schier A.F., Shendure J. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353:aaf7907. doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulco C.P., Munschauer M., Anyoha R., Munson G., Grossman S.R., Perez E.M., Kane M., Cleary B., Lander E.S., Engreitz J.M. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klann T.S., Black J.B., Chellappan M., Safi A., Song L., Hilton I.B., Crawford G.E., Reddy T.E., Gersbach C.A. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 2017;35:561–568. doi: 10.1038/nbt.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horlbeck M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A., Chen Y., Fields A.P., Park C.Y., Corn J.E., Kampmann M., Weissman J.S. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5:1–20. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid L.H., Gregg R.G., Smithies O., Koller B.H. Regulatory elements in the introns of the human HPRT gene are necessary for its expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1990;87:4299–4303. doi: 10.1073/pnas.87.11.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rincón-Limas D.E., Krueger D.A., Patel P.I. Functional characterization of the human hypoxanthine phosphoribosyltransferase gene promoter: evidence for a negative regulatory element. Mol. Cell. Biol. 1991;11:4157–4164. doi: 10.1128/mcb.11.8.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu S., Li W., Liu J., Chen C.H., Liao Q., Xu P., Xu H., Xiao T., Cao Z., Peng J. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diao Y., Fang R., Li B., Meng Z., Yu J., Qiu Y., Lin K.C., Huang H., Liu T., Marina R.J. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods. 2017;14:629–635. doi: 10.1038/nmeth.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zabidi M.A., Arnold C.D., Schernhuber K., Pagani M., Rath M., Frank O., Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesch M., Nyhan W.L.W. A familial disorder of uric acid metabolism and central nervous system function. Am. J. Med. 1964;36:561–570. doi: 10.1016/0002-9343(64)90104-4. [DOI] [PubMed] [Google Scholar]
- 34.Fu R., Ceballos-Picot I., Torres R.J., Larovere L.E., Yamada Y., Nguyen K.V., Hegde M., Visser J.E., Schretlen D.J., Nyhan W.L., Lesch-Nyhan Disease International Study Group Genotype-phenotype correlations in neurogenetics: Lesch-Nyhan disease as a model disorder. Brain. 2014;137:1282–1303. doi: 10.1093/brain/awt202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Essletzbichler P., Konopka T., Santoro F., Chen D., Gapp B.V., Kralovics R., Brummelkamp T.R., Nijman S.M., Bürckstümmer T. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aparicio-Prat E., Arnan C., Sala I., Bosch N., Guigó R., Johnson R. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics. 2015;16:846. doi: 10.1186/s12864-015-2086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs L., DeMars R. Chemical mutagenesis with diploid human fibroblasts. In: Kilbey B., Legator M., Nichols W., Ramcel C., editors. Handbook of Mutagenicity Test Procedures. Second Edition. Elsevier; 1984. pp. 321–356. [Google Scholar]
- 42.Monnat, R.J. (2009). Protocol for HPRT mutagenesis analyses. https://docs.wixstatic.com/ugd/944f87_bea96880f6ac402fb9199b6299f8163c.pdf.
- 43.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Döring A., Weese D., Rausch T., Reinert K. SeqAn an efficient, generic C++ library for sequence analysis. BMC Bioinformatics. 2008;9:11. doi: 10.1186/1471-2105-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 49.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 51.Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrne S.M., Ortiz L., Mali P., Aach J., Church G.M. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43:e21. doi: 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ardlie K.G., Deluca D.S., Segre A.V., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumasaka N., Knights A.J., Gaffney D.J. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanborn A.L., Rao S.S.P., Huang S.-C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patwardhan R.P., Lee C., Litvin O., Young D.L., Pe’er D., Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.