Abstract
Rationale: Heterogeneity in the septic response has hindered efforts to understand pathophysiology and develop targeted therapies. Source of infection, with different causative organisms and temporal changes, might influence this heterogeneity.
Objectives: To investigate individual and temporal variations in the transcriptomic response to sepsis due to fecal peritonitis, and to compare these with the same parameters in community-acquired pneumonia.
Methods: We performed genome-wide gene expression profiling in peripheral blood leukocytes of adult patients admitted to intensive care with sepsis due to fecal peritonitis (n = 117) or community-acquired pneumonia (n = 126), and of control subjects without sepsis (n = 10).
Measurements and Main Results: A substantial portion of the transcribed genome (18%) was differentially expressed compared with that of control subjects, independent of source of infection, with eukaryotic initiation factor 2 signaling being the most enriched canonical pathway. We identified two sepsis response signature (SRS) subgroups in fecal peritonitis associated with early mortality (P = 0.01; hazard ratio, 4.78). We defined gene sets predictive of SRS group, and serial sampling demonstrated that subgroup membership is dynamic during intensive care unit admission. We found that SRS is the major predictor of transcriptomic variation; a small number of genes (n = 263) were differentially regulated according to the source of infection, enriched for IFN signaling and antigen presentation. We define temporal changes in gene expression from disease onset involving phagosome formation as well as natural killer cell and IL-3 signaling.
Conclusions: The majority of the sepsis transcriptomic response is independent of the source of infection and includes signatures reflecting immune response state and prognosis. A modest number of genes show evidence of specificity. Our findings highlight opportunities for patient stratification and precision medicine in sepsis.
Keywords: gene expression, septic, immune, patient stratification
At a Glance Commentary
Scientific Knowledge on the Subject
There is significant heterogeneity in the septic response, which has hindered efforts to understand pathophysiology and develop targeted therapies. Molecular approaches may provide insights into variation in the host response, enabling biomarker development. Recent evidence suggests that transcriptomic sepsis response signatures (SRSs) can define patient subgroups associated with early outcome in sepsis due to community-acquired pneumonia (CAP).
What This Study Adds to the Field
This study provides the first substantive analysis of the transcriptomic response in patients admitted to intensive care with sepsis due to fecal peritonitis. Comparison with sepsis due to community-acquired pneumonia and with nonseptic control subjects demonstrated a shared sepsis response, independent of source of infection, that involved a significant proportion of the transcribed genome and overlapped with the “genomic storm” following trauma. We found evidence of SRS groups in patients with fecal peritonitis predictive of early mortality, with group membership changing over time in some patients. We show that the major predictor of variation in gene expression between patients with sepsis is SRS group rather than source of infection, with only a small number of genes differentially regulated according to the latter, enriched for IFN signaling and antigen presentation. These findings highlight opportunities for patient stratification in sepsis.
Sepsis is the life-threatening organ dysfunction caused by a dysregulated host response to infection (1). Therapeutic options remain limited despite extensive efforts to develop and refine treatment strategies (2). New insights into pathophysiology and the development of targeted treatments appropriate for individual patients at specific stages of the illness are urgently required (3). To be successful, this approach requires clearer understanding of heterogeneity in the sepsis response, in which proinflammatory and immunosuppressed states are dynamic and frequently coexist (4). We recently identified distinct transcriptomic sepsis response signatures (SRSs) in peripheral blood leukocytes from patients with community-acquired pneumonia (CAP) that are informative for immune response states and outcome (5). In particular, patients with SRS1 exhibit an immunosuppressed phenotype associated with higher early mortality, with features of endotoxin tolerance, T-cell exhaustion, and downregulation of HLA class II.
It is not yet known whether comparable SRSs are present in sepsis caused by other sources of infection. Additionally, the extent to which the source of infection contributes to heterogeneity in the transcriptomic response is unclear, although there is evidence that gene expression signatures can distinguish between gram-positive, gram-negative, and viral etiologies, and these signatures may be useful in the diagnosis of CAP (6–13). In fecal peritonitis (FP), sepsis is triggered by a polymicrobial infection within the peritoneal cavity, complicated by the release of damage-associated molecular patterns (DAMPs) and the effects of anesthesia (14). Conversely, CAP is caused by specific bacterial or atypical pathogens, and it is commonly preceded or caused by viral infection (15). Antibiotic choice, such as macrolides for CAP, may also have immunomodulatory effects (16, 17). Finally, whereas patients with FP usually have a rapid onset of illness, patients with CAP are often unwell for many days prior to intensive care unit (ICU) admission.
FP is a common cause of sepsis with a high mortality (18) in which longitudinal analysis is tractable, given that a defined time of onset can be estimated. We hypothesized that SRSs similar to those seen in CAP would be present in patients with FP, but that aspects of the transcriptomic host response would be dependent on the source of infection and stage of illness. In the present study, we investigated how patterns of gene expression in leukocytes are influenced by source of infection in FP and CAP, as well as how they vary between individual patients and within patients over time. Some of the results of these studies were previously reported in the form of abstracts (19, 20).
Methods
Study Design and Participants
The objective was to characterize the transcriptomic response to sepsis caused by FP, including an analysis of temporal changes and a comparison with sepsis caused by CAP. The subjects were adult patients admitted to the ICU with sepsis as part of the UK Genomic Advances in Sepsis (GAinS) study (www.ukccg-gains.org) with predefined inclusion and exclusion criteria (described in the online supplement). FP was diagnosed at laparotomy as inflammation of the peritoneal membrane secondary to large bowel perforation and fecal contamination (18). CAP was defined as a febrile illness associated with cough, sputum production, breathlessness, leukocytosis, and radiological features of pneumonia acquired in the community or within 2 days of ICU admission (21, 22).
This was a prospective observational study. The transcriptomic response was investigated in peripheral blood leukocytes and compared between patients and over time, and with control subjects undergoing elective cardiac surgery (described in the online supplement). Discovery and validation cohorts were used to identify shared and specific gene expression patterns among patients with FP and patients with CAP (see Figure E1 in the online supplement).
Sample Collection
Blood samples (5 ml) were collected (VACUETTE ethylenediaminetetraacetic acid–coated tubes; Greiner Bio-One, Kremsmünster, Austria) on the first, third, and/or fifth day after ICU admission. The total blood leukocyte population was isolated using LeukoLOCK filters (Life Technologies, Carlsbad, CA) and then stabilized using RNAlater (Life Technologies), followed by total RNA extraction (described in the online supplement). Extensive, anonymized clinical information was recorded using an electronic case report form (5).
Microarray Analysis
Genome-wide gene expression was quantified using Illumina HumanHT-12 v4 Expression BeadChips (47,231 probes; Illumina, San Diego, CA) with sample processing, data preparation, background subtraction, transformation, and normalization using the vsn package (23) (described in the online supplement). Gene expression data are available through the ArrayExpress database (accession number E-MTAB-5273/E-MTAB-5274).
Statistical Analysis
Analysis was performed using the R statistical software package (R Foundation for Statistical Computing, Vienna, Austria) unless otherwise specified, with statistical tests, power calculations, differential gene expression, enrichment testing, predictive modeling, and cluster analysis described in the online supplement.
Results
Transcriptomic Response to FP: Identification of SRSs
We quantified genome-wide gene expression for 117 adult patients with FP and 126 adult patients with CAP admitted to the ICU with sepsis, analyzing RNA from blood leukocytes rapidly isolated at the bedside. All patients showed evidence of organ dysfunction based on Sequential Organ Failure Assessment (SOFA) scores during ICU admission. These patients were recruited concurrently and processed in parallel as discovery (64 with FP, 73 with CAP) and validation cohorts (53 with FP, 53 with CAP; the 53 patients with CAP were previously published [5]). We also analyzed samples obtained from 10 subjects prior to elective cardiac surgery as nonseptic control subjects in the discovery cohort. Demographics and clinical covariates for the discovery set (FP, n = 64 patients; CAP, n = 73 patients; 221 samples; see Figure E1) are shown in Tables 1 and E1A.
Table 1.
Comparison of Clinical Characteristics of Patients with Sepsis, Based on Source of Infection in the Discovery and Validation Cohorts for the First Available Sample for Each Patient
| |
Discovery Cohort |
Validation Cohort |
|||||
|---|---|---|---|---|---|---|---|
| CAP (n = 73) | FP (n = 64) | P Value | CAP (n = 53) | FP (n = 53) | P Value | ||
| Age, yr |
60.6 (15.6) | 65.1 (17.5) | NS | 68.4 (13.7) | 68 (12.7) | NS | |
| Male sex |
37 (50.6%) | 28 (43.7%) | NS | 37 (69.8%) | 31 (58.5%) | NS* | |
| APACHE II score on Day 1 in the ICU |
18 (6.2) | 15 (5.9) | 0.005 | 19.7 (6.1) | 15.7 (5.9) | 0.0008 | |
| SOFA score on day of sampling |
6.8 (3.5) | 5.4 (4.0) | 0.03 | 6.2 (3.8) | 6.3 (3.5) | NS | |
| Mortality |
|||||||
| 14 d |
8 (11.0%) | 6 (9.4%) | NS† | 7 (13.2%) | 6 (11.3%) | NS† | |
| 28 d |
13 (17.8) | 8 (12.5%) | NS† | 7 (13.2%) | 7 (13.2%) | NS† | |
| 6 mo |
17 (23.3%) | 11 (17.2%) | NS† | 11 (20.8%) | 12 (22.6%) | NS† | |
| Infection | |||||||
| Gram-positive bacteria |
14 (19.2%) | 7 (13.2%) | |||||
| Gram-negative bacteria |
8 (11.0%) | 5 (9.4%) | |||||
| Viral |
6 (8.2%) | 5 (9.4%) | |||||
| Mechanical ventilation |
51 (69.9%) | 34 (53.1%) | 0.05 | 19 (35.8%) | 21 (39.6%) | NS* | |
| Respiratory rate, breaths/min |
29.2 (9.2) | 20.0 (8.2) | <0.0001 | 31.1 (8.7) | 22.7 (9.0) | <0.0001 | |
| Days of respiratory support |
9.6 (13.5) | 4.7 (8.1) | 0.01 | 8.8 (12.1) | 9.3 (14.8) | NS | |
| Oxygenation index |
19.3 (9.9) | 30.9 (12.3) | <0.0001 | 21.2 (7.5) | 28.2 (12.2) | <0.0001 | |
| Vasopressors/inotropes |
NS | NS* | |||||
| No dose |
37 (50.7%) | 35 (54.7%) | 36 (67.9%) | 27 (50.9%) | |||
| Low dose |
1 (1.4%) | 1 (1.6%) | 0 (0%) | 0 (0%) | |||
| Medium dose |
10 (13.7%) | 5 (7.8%) | 7 (13.2%) | 6 (11.3%) | |||
| High dose |
24 (32.9%) | 23 (35.9%) | 10 (18.9%) | 20 (37.7%) | |||
| Mean arterial pressure, lowest, mm Hg |
65.0 (12.8) | 67.7 (12.5) | NS | 69.6 (11.1) | 67.5 (14.6) | NS | |
| Temperature, low, °C |
36.4 (0.8) | 36.1 (0.7) | 0.018 | 36.0 (1.0) | 36.1 (0.6) | NS | |
| Hematocrit, % |
35.4 (6.9) | 30 (5.9) | <0.0001 | 35.0 (7.9) | 31.5 (6.6) | 0.02 | |
| Proportion of leukocytes |
|||||||
| Lymphocytes |
9.17% | 6.52% | 0.008 | 9.05% | 8.77% | NS | |
| Polynucleocytes |
84.10% | 87.98% | 0.029 | 83.40% | 85.30% | NS | |
| Mononucleocytes |
6.73% | 5.50% | NS | 7.51% | 5.95% | NS | |
| Bicarbonate, mmol/L |
25.0 (6.8) | 22.1 (5.1) | 0.008 | 24.6 (6.8) | 23.1 (5.0) | NS | |
| Renal replacement therapy | 6 (8.2%) | 5 (7.8%) | NS | 5 (9.4%) | 8 (15.1%) | NS* | |
Definition of abbreviations: APACHE II = Acute Physiology and Chronic Healthy Evaluation II; CAP = community-acquired pneumonia; FP = fecal peritonitis; ICU = intensive care unit; NS = not significant; SOFA = Sequential Organ Failure Assessment.
Data are count (percent) or mean (SD) unless otherwise specified. Statistical analysis was done by t test unless otherwise specified.
Chi-square test.
Log-rank test.
We investigated whether sepsis response subgroups based on global patterns of gene expression were present in FP. The combined FP cohort (147 samples from 117 patients) had a 28-day mortality of 13%; the mean age was 66 years, and 50.4% of patients were male (Table 2, Table E1B). We applied two complementary approaches to this FP cohort: (1) hypothesis-free unsupervised hierarchical clustering based on observed patterns of variation in gene expression and (2) assignment based on expression of a specific seven-gene set previously shown to be predictive of SRS group membership in CAP (5).
Table 2.
Characteristics of Patients with Fecal Peritonitis (All Patients) on Day 1
| Characteristics | Data |
|---|---|
| Number of patients | 117 |
| Age, yr, mean ± SD | 66.4 (±15.5) |
| Male sex, % | 50.4% |
| Median (range) APACHE II score | 15 (2–40) |
| Median (range) SOFA score on day of sampling | 5 (0–18) |
| Mortality | |
| 14 d | 12 (10.3%) |
| 28 d | 15 (12.8%) |
| 6 mo | 23 (19.7%) |
| Cause of peritonitis | |
| Diverticular disease | 30 (25.6%) |
| Surgical anastomosis breakdown | 28 (23.9%) |
| Malignancy | 14 (12%) |
| Trauma | 3 (2.6%) |
| Other/unknown | 42 (35.9%) |
| Mechanical ventilation | 55 (47%) |
| Vasopressors | 55 (47%) |
| Renal replacement therapy | 13 (11%) |
| Mean (SD) arterial pressure, lowest, mm Hg | 68.2 (14.8) |
Definition of abbreviations: APACHE II = Acute Physiology and Chronic Healthy Evaluation II; SOFA = Sequential Organ Failure Assessment.
Data are count (percent) unless otherwise specified.
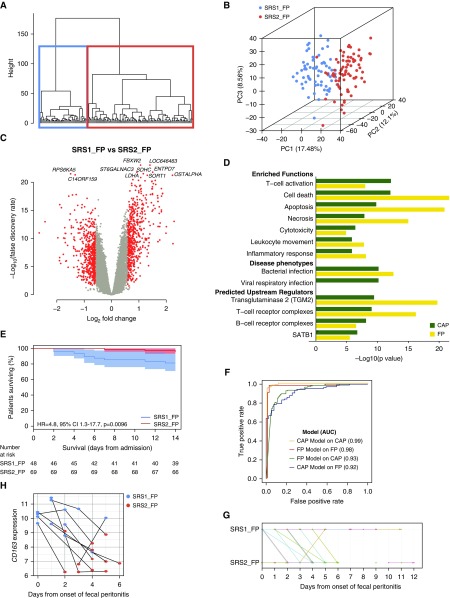
We first analyzed variation in global gene expression (2,716 probes representing the top 10% most variably expressed genes) using agglomerative hierarchical clustering followed by consolidation of group membership using k-means. This approach revealed two distinct patient groups: SRS1_FP (n = 68 [46%]) and SRS2_FP (n = 79 [54%]) (Figures 1A and 1B). Following group assignment, 1,075 genes were found to be differentially expressed between these groups (Figure 1C, Table E1C), showing strong correlation with those previously observed in patients with CAP (5) (Pearson’s r = 0.791) (Figure E2). To determine the functional overlap with SRS groups identified in CAP, we performed pathway, function enrichment, and upstream regulator analysis (Figure 1D, Table E1D). Cell death, apoptosis, necrosis, T-cell activation, and endotoxin tolerance were significantly enriched biological functions in SRS1 for both sources of infection. We proceeded to test for association with outcome in FP and found that SRS1_FP group membership was associated with higher early mortality (14-d mortality log-rank test P = 0.0096; 18.8% vs. 4.3%; hazard ratio, 4.78; 95% confidence interval, 1.29–17.65) (Figure 1E). The effect of SRS group membership remained after inclusion of age, SOFA score, Charlson comorbidity index, and sex in a Cox proportional hazards model (P = 0.037; hazard ratio, 4.23; 95% confidence interval, 1.09–16.39).
Figure 1.
Transcriptomic sepsis response signatures (SRSs) in fecal peritonitis (FP). (A) Unsupervised hierarchical clustering analysis of 147 FP samples for the top 10% most variable probes (n = 2,716). (B) First three principal components (PCs) plotted with the proportion of variance explained by each component, with samples colored by SRS group membership assigned by hierarchical and k-means clustering. (C) Volcano plot of differentially expressed probes for SRS1_FP versus SRS2_FP (red coloring shows fold change >1.5, false discovery rate <0.05). (D) Enriched functions, disease phenotypes, and predicted upstream regulators derived from differentially expressed probes in FP SRS groups and compared with the previously published community-acquired pneumonia (CAP) dataset. Enrichment was also seen for an endotoxin tolerance gene expression signature we previously defined (5) using publicly available datasets (42, 43) in SRS1_FP relative to SRS2_FP, tested using ROAST (rotation gene set testing), a gene set enrichment test (P < 1 × 10−5). (E) Kaplan-Meier survival plot by SRS group (shaded areas, 95% confidence intervals [CIs]) with a single sample selected at random for those patients with multiple samples to assign SRS membership. (F) The performance of the SRS group assignment models (gene sets) which were derived and tested in the FP and previously published CAP datasets, are shown by receiver operating characteristic curves, and the area under the curve (AUC) is given for each. (G) Time course of patient SRS-FP group membership using serial samples and days from disease onset. (H) Expression of CD163 over time from disease onset in samples from the 11 patients with FP who moved between SRS groups. Each point represents a sample, colored according to SRS group assignment, with lines linking samples from the same patient. HR = hazard ratio.
To further validate our findings, we adopted a second approach, namely assignment of SRS group membership in this FP cohort using the seven-gene set (DYRK2, CCNB1IP1, TDRD9, ZAP70, ARL14EP, MDC1, and ADGRE3) we previously established in patients with CAP (5) (Figure E3). This showed strong concordance with the results obtained by unsupervised analysis (misclassification rate, 21.1%; area under the curve [AUC], 0.923) (Figure 1F). The groups defined using the seven-gene set showed a significant difference in early mortality rates (log-rank test P = 0.030 for patients with FP), a differential gene expression signature strongly correlated with that seen in the original CAP analysis (r = 0.845), as well as pathway enrichment comparable to findings in patients with CAP. We also derived a six-gene set (CD163, ZDHHC19, MME, FAM89A, ZBP1, and B3GNT2) from the FP data predictive of SRS_FP group membership, with a 4.1% misclassification rate based on internal leave-one-out cross-validation (AUC, 0.975) (Figure 1F). When applied to the original CAP cohort (5), this six-gene set again performed well (misclassification rate, 27.9%; AUC, 0.931).
A poor outcome subgroup has previously been reported in children with sepsis (24). We investigated the similarity between pediatric endotype A and SRS1 by comparative differential gene expression analysis. Although there was some overlap in pathway enrichment (e.g., T- and B-cell receptor signaling), the gene expression patterns that distinguish SRS groups were not enriched in the pediatric endotype contrast (Figure E4).
Temporal Changes in SRSs
Serial sampling on Day 1, Day 3, and Day 5 following ICU admission provides the opportunity to investigate the relationship between SRS group membership and disease progression. We found that 11 (46%) of 24 patients with FP with serial samples moved between groups over time; 10 moved from SRS1 to SRS2, only 1 of whom died (Figure 1G). Thirteen patients (54%) remained in the same SRS group; three of seven patients remaining in SRS1 died, compared with no deaths among the six remaining in SRS2. This movement between SRS groups involves large changes in gene expression, illustrated by CD163, encoding an innate immune sensor for bacteria, and one of the six gene classifiers for SRS_FP (Figures 1H and E5).
Influence of Source of Infection on the Transcriptomic Response
We proceeded to further characterize and compare gene expression in patients with FP and patients with CAP to determine the relative importance of shared and specific features of the transcriptomic response. As expected, in the discovery cohort, gastrointestinal comorbidities were more common in patients with FP, whereas patients with CAP had more respiratory comorbidities, higher respiratory rates and oxygenation requirements, and higher lymphocyte counts (Table 1 and Table E1A). Although mortality did not differ significantly, patients with CAP had higher Acute Physiology and Chronic Healthy Evaluation II and SOFA scores than patients with FP (Table 1).
To understand the relationships between source of infection, SRS group, and heterogeneity in the septic response, we performed principal component analysis using the top 10% most variably expressed transcripts. Nonseptic control subjects clustered together and were clearly distinct from septic patients (Figure 2A). Among the combined population of patients with sepsis, there was clear segregation based on SRS group but not source of infection (Figure 2A).
Figure 2.
Transcriptomic response to sepsis. (A) First two principal components (PCs) of gene expression data plotted with the proportion of variance explained by each component shown. Solid circles represent fecal peritonitis (FP) discovery samples (n = 94), and open circles represent community-acquired pneumonia (CAP) discovery samples (n = 127). Samples are colored according to sepsis response signature (SRS) group assignment, showing that sepsis response states have considerable overlap between sources of infection. Control subjects (n = 10) are indicated by x. (B) Heatmap showing correlation between the first six PCs, SRSs, and clinical covariates for sepsis samples (FP, n = 94; CAP, n = 127). (C) Venn diagram showing the overlap in differential gene expression versus control subjects in the sepsis response and the response to trauma (false discovery rate [FDR], <0.05; fold change [FC], >1.5; first 5 d). Selected condition-specific enriched pathways and biological functions are noted. (D) Volcano plot of differentially expressed probes for FP (n = 94; left) and CAP (n = 127; right) versus control subjects (n = 10) (red coloring shows FC >1.5, FDR <0.05). APACHE = Acute Physiology and Chronic Health Evaluation; EIF2 = eukaryotic initiation factor 2; NFAT = nuclear factor of activated T cells; SOFA = Sequential Organ Failure Assessment; TNFR1 = tumor necrosis factor receptor 1.
To further elucidate the drivers of variation in the septic response, we calculated correlations between the observed variance in global gene expression for the sepsis samples (the first six principal components of variance, representing approximately 50% of the variance in the data), a comprehensive set of clinical covariates, and SRS group (Figure 2B, Table E1E). This showed that variation in gene expression was most strongly correlated with SRS group membership (PC1 r = 0.77; false discovery rate [FDR], <2.2 × 10−16). More modest correlation was seen with source of infection (PC1 r = −0.417; FDR, 6.12 × 10−9), bicarbonate levels (PC1 r = 0.35; FDR, 2.2 × 10−8), and neutrophil count (PC1 r = −0.29; FDR, 2.0 × 10−3). When the dataset was divided into CAP and FP samples and the analysis repeated, we found that SRS remained the most significant correlate (Figure E6).
When we compared all patients in the discovery cohort (FP, n = 64; CAP, n = 73) with nonseptic control subjects (n = 10), we found that sepsis was associated with differential regulation of a large proportion of the transcribed genome (18.0% of transcripts assayed; 4,881 probes; >1.5-fold change [FC]; FDR, <0.05) (Table E1F). Pathway analysis showed that eukaryotic initiation factor 2 signaling was the top canonical pathway, consistent with the reported role for this translational initiation factor in response to viral and bacterial infection (25), and predicted key upstream regulators IL-13, ATB1, TGFB1, and IL-2 (Table E1G). We used the same methodology to compare gene expression in trauma patients with healthy subjects, using the previously published genomic response to trauma dataset (26) but restricting this to samples from the same time frame as the sepsis cohort (5 d following injury). We found that, of the genes differentially expressed in the sepsis response and measured in the trauma cohort, the majority (n = 1,884 [64%]) were also differentially expressed in trauma (Figure 2C). Commonality was seen with eukaryotic initiation factor 2 signaling and inflammation-related pathways enriched in both sepsis and trauma responses, whereas the inflammatory response to organismal injury was specific to trauma and tumor necrosis factor receptor 1 (TNFR1) signaling was specific to sepsis (Figure 2C, Table E1H).
When we compared patients with CAP versus control subjects and patients with FP versus control subjects, we observed that gene expression patterns were highly correlated, demonstrating that most sepsis response pathways are common and independent of source of infection (Figures 2D, 3A, and 3B; Tables E1I and E1J). To further investigate differential gene expression between patients with sepsis due to FP or CAP, we compared the first available samples for each patient following ICU admission (FP, n = 64; CAP, n = 73). We found 310 probes (263 genes) differentially expressed between the two sources of infection (FC, >1.5; FDR, <0.05) (Figure 3C, Table E1K), significantly more than expected by chance based on permutation analysis (Figure E7). We noted significant differences in total and differential cell counts between CAP and FP. The inclusion of differential cell counts in the regression model had minimal influence, with a similar list of differentially regulated genes and FCs strongly correlated with the original analysis (Figure E8, Table E1K). IFN signaling (P = 6.1 × 10−10) and antigen presentation (P = 1.6 × 10−8) were the most significant enriched canonical pathways, upregulated in patients with CAP compared with FP (Figure 3D, Table E1L). Enriched networks involved IFN-α/β and the antimicrobial/inflammatory response (P = 1 × 10−39) (Figure 3E). IFIT1, IFIT2, and IFIT3 (IFN-induced antiviral genes), BTN3A3 (major histocompatibility complex class I gene), IFIH1 (sensor of viral nucleic acids), and OAS2 (IFN-induced antiviral enzyme) were all upregulated in patients with CAP. IFNs, endotoxins, and TNF were found to be significant upstream regulators (Figure 3D, Table E1L).
Figure 3.
Variation in the sepsis transcriptome according to source of infection. (A) Venn diagram showing the overlap in the fecal peritonitis (FP) and community-acquired pneumonia (CAP) sepsis response in terms of the number of differentially expressed genes versus control subjects (FP, n = 64 samples; CAP, n = 73 samples; control subjects, n = 10 samples). (B) Correlation of differential gene expression (DE) between sepsis due to FP (n = 64 samples) and control subjects (n = 10 samples), and sepsis due to CAP (n = 73 samples) and control subjects (n = 10 samples). (C) Volcano plot of differentially expressed probes for CAP versus FP (red coloring shows fold change [FC] >1.5, FDR <0.05; positive fold change indicates relative upregulation in CAP). (D) Enriched pathways and predicted upstream regulators derived from differentially expressed genes for CAP versus FP in the discovery (CAP, n = 73 samples; FP, n = 64 samples) and validation cohorts (CAP, n = 53 samples; FP, n = 53 samples). (E) Most significantly enriched network (P = 1 × 10−39), based on comparison of CAP versus FP with differentially expressed genes shaded (red = upregulation; green = downregulation), and logarithmic FC with P value shown for each gene. (F) Volcano plot of the differentially expressed probes for positive viral diagnosis (n = 25) versus no positive viral diagnosis (n = 240) in 265 patients with CAP. Red coloring indicates FC less than 1.5, FDR less than 0.05. IRF = IFN regulatory factor; MHC = major histocompatibility complex; STAT = signal transducer and activator of transcription; TNF = tumor necrosis factor.
We confirmed our findings in a validation set comprising 53 patients with FP and 53 patients with CAP prospectively recruited to the UK GAinS study (Figure E1). We found SRS group, source of infection, cell count, and day of sampling were strongly associated with the first six principal components of gene expression; genes differentially expressed between FP/CAP correlated with those found in the discovery cohort (Pearson’s r = 0.83; P < 2.2 × 10−16) (Table E1M). Pathway analysis demonstrated high concordance of enriched pathways and functions (Figure 3D, Table E1N). We found that a gene set comprising EPHB1, NQO2, ARG1, HMGB2, FGL2, and GPR162 was predictive of source of infection (FP or CAP) for the discovery cohort. We applied a sparse regression method to show that, for the discovery cohort, the misclassification rate was 29.9% with internal leave-one-out validation (AUC, 0.760); for the validation cohort, the test error was 28.3% (AUC, 0.798) (Figure E9). There was no difference in the proportion of different sampling days following ICU admission between FP and CAP samples in the analyzed cohorts (chi-square P = 0.350) (Figure E10).
Given a prominent viral signature among genes differentially expressed between patients with FP and patients with CAP, we investigated if this came from a subset of patients with CAP with a viral infection. Only 6 of 73 patients with CAP in the discovery cohort had a confirmed viral infection; we therefore analyzed the transcriptomic differences between patients with and without confirmed viral infection in our larger, previously published CAP cohort (25 vs. 240 patients) (5). We identified 39 differentially expressed genes (FDR, <0.05; FC, >1.5) (Figure 3F, Table E1O), including IFI27, an IFN-α–inducible protein reported as a marker of influenza infection (27), and LAMP3, a dendritic cell glycoprotein induced by influenza A infection (28). Pathway analysis showed the most significant enrichment for pattern recognition receptors, IFN signaling, and IFN regulatory factors (Table E1P). We then compared this gene set to the genes differentially expressed between patients with FP and patients with CAP and found significant enrichment of 24 of 39 genes (P < 1 × 10−4).
Temporal Changes in Gene Expression
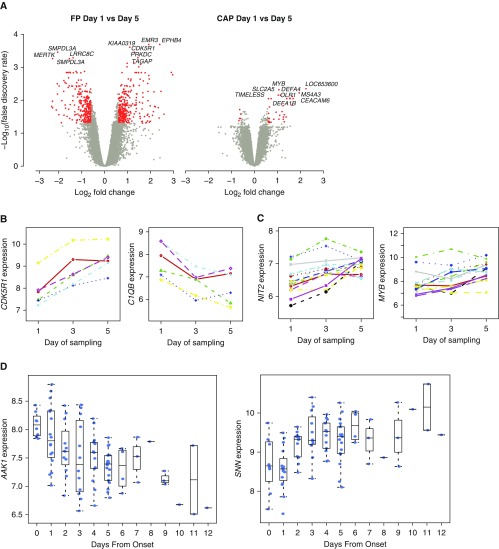
Interpretation of gene expression patterns in sepsis may be complicated by dynamic differences in the host response over time (10, 29). We explored temporal changes in gene expression for patients with FP and patients with CAP from whom serial samples following ICU admission were available. We first sought to identify genes that varied over time using a repeated measures regression model. We found that 714 genes were significantly differentially expressed in patients with FP sepsis between Days 1 and 5, compared with only 80 genes in patients with CAP (FDR, <0.05; FC, >1.5) (Figure 4A, Table E1Q). We further analyzed our data using a multivariate empirical Bayes model that ranks genes based on differential expression for longitudinal series involving multiple biological conditions (30), restricting this analysis to patients from whom samples were available at all three time points. Notably, temporal changes in gene expression were again more pronounced in patients with FP (Table E1R), and the specific genes whose expression changed over time differed between FP and CAP (Figures 4B and C).
Figure 4.
Dynamics of gene expression in sepsis due to fecal peritonitis (FP) and community-acquired pneumonia (CAP). (A) Volcano plot of the differentially expressed probes for Day 1 versus Day 5 samples from the same patient in FP (n = 9; left) and CAP (right; n = 17) (red coloring indicates fold change >1.5 and false discovery rate <0.05). (B) Examples of genes showing significant temporal variation in expression over time for patients with sepsis due to FP include the kinase activator CDK5R1 and complement component C1QB; other examples are the adhesion receptor EMR3 and cysteine protease CAPN13. (C) Examples of genes showing variation in expression over time for patients with sepsis due to CAP include cell cycle gene NIT2 and the hematopoiesis regulatory transcription factor gene MYB; other examples are the antimicrobial neutrophil peptidase CTSG, inflammatory regulator CAMP, defensin DEFA4, and neutrophil granule elastase ELANE. (D) Further examples of genes differentially expressed in FP over time plotted from time of onset of FP include the adaptor-related protein complex 2–associated kinase 1 (AAK1), involved in regulating clathrin-mediated endocytosis (44), which is critical for bacterial entry (45), as well as SNN encoding stannin, a mitochondrial damage sensor (46).
In patients with FP, it was possible to estimate with reasonable accuracy the time of onset of the acute event. We calculated days from disease onset for each FP sample and used this to investigate temporal changes. Using a linear model with limma analysis, we found that 140 genes showed significant changes in expression over time (FDR, <0.05) (Table E1S), including AAK1 and SNN (Figure 4D). Pathway analysis was significant for genes involved in phagosome formation; natural killer cell and IL-3 signaling with evidence of enrichment for specific biological functions, notably cellular degranulation, chemotaxis, leukocyte activation, macrophage adhesion, phagocytosis, and bacterial infection (Table E1T).
Discussion
We have characterized the transcriptomic response to sepsis caused by FP and found evidence of SRSs associated with outcome, with patients in SRS1 having higher early mortality. Transcriptomic features of endotoxin tolerance and pathway enrichment for cell death, apoptosis, necrosis, and T-cell exhaustion are consistent with animal models and human studies demonstrating the importance of immune compromise in sepsis pathogenesis and as a determinant of poor outcomes (4, 31, 32). The FP dataset allowed us to explore evolution of SRS membership. We found a significant number of patients (46%) switched SRS groups in the first 5 days of ICU admission, with the majority moving from SRS1 to SRS2. Persistence of SRS1 is associated with a poor outcome, whereas maintenance or recovery of immune competence (SRS2) is associated with survival. These findings further support the concept that SRS group membership reflects clinically important biological differences (5) and suggest that, if used as a biomarker, the transcriptomic response signature should be determined at the time a therapeutic intervention is being considered. Establishing the immune response state of a patient could enable individually tailored immunotherapy and monitoring of the response to treatment.
We found that the transcriptomic response is, to a large extent, shared between the two sources of infection we analyzed, with the expression of only a modest number of genes being dependent on source of infection. This shared sepsis response involves a significant proportion of the transcribed genome and overlapped with the “genomic storm” following trauma, although we observed some differences involving, for example, TNFR1 signaling (26). Regarding gene expression between FP and CAP, the most enriched networks involved IFN-α/β and the antimicrobial/inflammatory response. These differences seem to be driven predominantly by viral respiratory infection within the CAP cohort; however, given current difficulties in pathogen detection, the biological and clinical interpretation of such transcriptomic differences remains challenging. Whereas some previous studies have supported a common transcriptional septic response independent of pathogen (33), others have reported that expression signatures can discriminate between infecting organisms (6–8, 13), although these findings remain controversial (9, 10). For patients admitted to the ICU with suspected CAP, a 78-transcript signature has been reported to differentiate cases of CAP from non-CAP, with the FAIM3:PLAC8 gene expression ratio proposed as a diagnostic biomarker (11).
We found that temporal changes were more pronounced in patients with FP, with more than eight times as many genes differentially expressed between admission and Day 5 than in patients with CAP, and these changes involved phagosome formation, natural killer cell signaling, IL-3 signaling, leukocyte activation, mitochondrial damage, and apoptosis. To date, time series analyzing changes in gene expression have been focused on animal models of sepsis (34), the endotoxin response in healthy volunteers (35), the response to trauma and subsequent infection (26), and early events in sepsis (29), and authors of a recent analysis of time-matched cohorts defined a gene set distinguishing sterile inflammation from infectious inflammation (12). In general, clearer resolution of temporal differences is critical to resolving heterogeneity in observed sepsis responses within and between patients.
Future work should include comprehensive pathogen phenotyping, such as by using metagenomic sequencing, given the recognized high proportion of culture-negative patients with sepsis (36, 37). Comparison of patients with FP with specific control subjects, such as a group of patients undergoing laparotomy for noninfectious indications, would control for the effects of damage-associated molecular pattern–mediated signaling due to the surgical procedure as well as any modulating influences on the transcriptome due to general anesthesia, whereas inclusion of patients undergoing laparotomy for gastric or small bowel perforation would to some extent control for different pathogens. Differences in differential cell count are potential confounders in transcriptomic analysis (10), and, although the results presented were robust to differential leukocyte count, cell-type–specific transcriptomic profiling will be important for future studies (38).
Defining the most robust and informative predictive gene set for SRS membership on the basis of transcriptomics requires prospective large-scale validation using an appropriate point-of-care test. We note that the current seven-gene set established in patients with CAP and validated in patients with FP is consistent with a key role for a dysfunctional immune response in sepsis. This predictive gene set includes genes involved in cell growth (DYRK2), cell cycle (CCNB1IP1), stem cell maintenance (TDRD9), and DNA damage (ADGRE3), consistent with the observed pathway enrichment (cell death, apoptosis, necrosis) and with immune response through lymphocyte activation (ZAP70), major histocompatibility complex class II export (ARL14EP), and myeloid cell interactions in immunity (MDC1). The lack of overlap with pediatric sepsis endotypes that we observed requires further validation and may reflect developmental differences impacting pathophysiology and host immune dysfunction (39, 40).
We have addressed sepsis heterogeneity using a transcriptomic approach in patients with FP and patients with CAP, demonstrating a shared sepsis response with distinct SRS groups that are dynamic, reflect the underlying biological response, and are informative for prognosis. Our findings also provide some evidence for differential patterns of expression between CAP and FP, two of the most common causes of sepsis. Our analysis, coupled with recent advances in the field (3, 4, 11, 41), highlights the opportunity to develop novel therapeutic interventions that can be targeted and appropriately timed to individual patients on the basis of their transcriptomic signatures, providing opportunities for precision medicine in sepsis.
Acknowledgments
Acknowledgment
The authors thank all participating patients, GAinS investigators, and recruiting hospitals (Table E2).
Footnotes
Supported by the National Institute for Health Research (NIHR) through the Comprehensive Clinical Research Network for patient recruitment, the Wellcome Trust (grants 074318 [J.C.K.] and 090532/Z/09/Z [core facilities Wellcome Trust Centre for Human Genetics including High-Throughput Genomics Group]), the European Research Council (ERC) under the European Union’s Seventh Framework Program (FP7/2007–2013)/ERC grant agreement 281824 (J.C.K.), the Medical Research Council (98082 [J.C.K.]), the UK Intensive Care Society, and the NIHR Oxford Biomedical Research Centre. A.V.S.H. is supported by a Wellcome Trust Senior Investigator Award (HCUZZ0), and A.C.G. is supported by an NIHR Clinician Scientist Fellowship. The organizations funding this study had no role in the design or conduct of the study; in the collection, management, analysis, or interpretation of the data; or in the preparation, review, or approval of the manuscript.
Author Contributions: J.C.K. and C.J.H.: conceived of the study; K.L.B., E.E.D., J.R., A.C.G., J.C.K., and C.J.H.: contributed to the study concept and design; K.L.B., E.E.D., J.R., and E.S.-J.: performed the laboratory work; K.L.B., E.E.D., J.R., P. Humburg, and J.C.K.: contributed to the statistical analysis; P. Hutton, C.G., and A.V.S.H.: provided administrative, technical, or material support; J.C.K., K.L.B., E.E.D., and C.J.H.: contributed to drafting of the manuscript; J.C.K. and C.J.H.: were responsible for obtaining funding; J.C.K.: led the study; and all authors: participated in the acquisition, analysis, or interpretation of data and also revised and approved the final version of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201608-1685OC on December 30, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20:195–203. doi: 10.1016/j.molmed.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J, Vincent JL, Adhikari NK, Machado FR, Angus DC, Calandra T, Jaton K, Giulieri S, Delaloye J, Opal S, et al. Sepsis: a roadmap for future research. Lancet Infect Dis. 2015;15:581–614. doi: 10.1016/S1473-3099(15)70112-X. [DOI] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, Rautanen A, Gordon AC, Garrard C, Hill AVS, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4:259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu SL, Chen HW, Yang PC, Peck K, Tsai MH, Chen JJ, Lin FY. Differential gene expression in gram-negative and gram-positive sepsis. Am J Respir Crit Care Med. 2004;169:1135–1143. doi: 10.1164/rccm.200211-1278OC. [DOI] [PubMed] [Google Scholar]
- 8.Feezor RJ, Oberholzer C, Baker HV, Novick D, Rubinstein M, Moldawer LL, Pribble J, Souza S, Dinarello CA, Ertel W, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun. 2003;71:5803–5813. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang BM, McLean AS, Dawes IW, Huang SJ, Cowley MJ, Lin RC. Gene-expression profiling of gram-positive and gram-negative sepsis in critically ill patients. Crit Care Med. 2008;36:1125–1128. doi: 10.1097/CCM.0b013e3181692c0b. [DOI] [PubMed] [Google Scholar]
- 10.Maslove DM, Wong HR. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scicluna BP, Klein Klouwenberg PM, van Vught LA, Wiewel MA, Ong DS, Zwinderman AH, Franitza M, Toliat MR, Nürnberg P, Hoogendijk AJ, et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J Respir Crit Care Med. 2015;192:826–835. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 12.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra71. doi: 10.1126/scitranslmed.aaa5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sweeney TE, Wong HR, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med. 2016;8:346ra91. doi: 10.1126/scitranslmed.aaf7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krepel CJ, Gohr CM, Edmiston CE, Condon RE. Surgical sepsis: constancy of antibiotic susceptibility of causative organisms. Surgery. 1995;117:505–509. doi: 10.1016/s0039-6060(05)80249-1. [DOI] [PubMed] [Google Scholar]
- 15.Marik PE Norasept II Study Investigators. The clinical features of severe community-acquired pneumonia presenting as septic shock. J Crit Care. 2000;15:85–90. doi: 10.1053/jcrc.2000.16460. [DOI] [PubMed] [Google Scholar]
- 16.Weiss T, Shalit I, Blau H, Werber S, Halperin D, Levitov A, Fabian I. Anti-inflammatory effects of moxifloxacin on activated human monocytic cells: inhibition of NF-κB and mitogen-activated protein kinase activation and of synthesis of proinflammatory cytokines. Antimicrob Agents Chemother. 2004;48:1974–1982. doi: 10.1128/AAC.48.6.1974-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovaleva A, Remmelts HH, Rijkers GT, Hoepelman AI, Biesma DH, Oosterheert JJ. Immunomodulatory effects of macrolides during community-acquired pneumonia: a literature review. J Antimicrob Chemother. 2012;67:530–540. doi: 10.1093/jac/dkr520. [DOI] [PubMed] [Google Scholar]
- 18.Tridente A, Clarke GM, Walden A, McKechnie S, Hutton P, Mills GH, Gordon AC, Holloway PA, Chiche JD, Bion J, et al. GenOSept Investigators. Patients with faecal peritonitis admitted to European intensive care units: an epidemiological survey of the GenOSept cohort. Intensive Care Med. 2014;40:202–210. doi: 10.1007/s00134-013-3158-7. [DOI] [PubMed] [Google Scholar]
- 19.Burnham K, Davenport E, Radhakrishnan J, Svoren-Jabalera E, Humburg P, Hutton P, Gordon A, Hill A, Hinds C, Knight J. Shared and specific features of the individual transcriptomic response in sepsis due to faecal peritonitis. Poster presented at the American Society of Human Genetics 2016 annual meeting. October 18–22, 2016, Vancouver, BC, Canada. Poster 3316W. [Google Scholar]
- 20.Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Hutton P, Garrard C, Hinds CJ, Knight JC. Heterogeneity in the individual transcriptomic response to severe sepsis. Presented at the 13th International Congress of Human Genetics. April 3–7, 2016, Kyoto, Japan. [Google Scholar]
- 21.Angus DC, Marrie TJ, Obrosky DS, Clermont G, Dremsizov TT, Coley C, Fine MJ, Singer DE, Kapoor WN. Severe community-acquired pneumonia: use of intensive care services and evaluation of American and British Thoracic Society Diagnostic criteria. Am J Respir Crit Care Med. 2002;166:717–723. doi: 10.1164/rccm.2102084. [DOI] [PubMed] [Google Scholar]
- 22.Walden AP, Clarke GM, McKechnie S, Hutton P, Gordon AC, Rello J, Chiche JD, Stueber F, Garrard CS, Hinds CJ ESICM/ECCRN GenOSept Investigators. Patients with community acquired pneumonia admitted to European intensive care units: an epidemiological survey of the GenOSept cohort. Crit Care. 2014;18:R58. doi: 10.1186/cc13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 24.Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shrestha N, Bahnan W, Wiley DJ, Barber G, Fields KA, Schesser K. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J Biol Chem. 2012;287:28738–28744. doi: 10.1074/jbc.M112.375915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, Ramilo O, Mejias A, Flaño E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol. 2012;86:5422–5436. doi: 10.1128/JVI.06757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, Xue Q, Wan Y, Yang Y, Wang J, Hung T. Lysosome-associated membrane glycoprotein 3 is involved in influenza A virus replication in human lung epithelial (A549) cells. Virol J. 2011;8:384. doi: 10.1186/1743-422X-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cazalis MA, Lepape A, Venet F, Frager F, Mougin B, Vallin H, Paye M, Pachot A, Monneret G. Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive Care Med Exp. 2014;2:20. doi: 10.1186/s40635-014-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai YC, Speed TP. On gene ranking using replicated microarray time course data. Biometrics. 2009;65:40–51. doi: 10.1111/j.1541-0420.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- 31.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD, II, Kreisel D, Krupnick AS, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 2011;306:2594–2605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pena OM, Hancock DG, Lyle NH, Linder A, Russell JA, Xia J, Fjell CD, Boyd JH, Hancock RE. An endotoxin tolerance signature predicts sepsis and organ dysfunction at initial clinical presentation. EBioMedicine. 2014;1:64–71. doi: 10.1016/j.ebiom.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 34.Mira JC, Szpila BE, Nacionales DC, Lopez MC, Gentile LF, Mathias BJ, Vanzant EL, Ungaro R, Holden D, Rosenthal MD, et al. Patterns of gene expression among murine models of hemorrhagic shock/trauma and sepsis. Physiol Genomics. 2016;48:135–144. doi: 10.1152/physiolgenomics.00072.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talwar S, Munson PJ, Barb J, Fiuza C, Cintron AP, Logun C, Tropea M, Khan S, Reda D, Shelhamer JH, et al. Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol Genomics. 2006;25:203–215. doi: 10.1152/physiolgenomics.00192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phua J, Ngerng W, See K, Tay C, Kiong T, Lim H, Chew M, Yip H, Tan A, Khalizah H, et al. Characteristics and outcomes of culture-negative versus culture-positive severe sepsis. Crit Care. 2013;17:R202. doi: 10.1186/cc12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, et al. EPIC II Group of Investigators. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 38.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aneja R, Carcillo J. Differences between adult and pediatric septic shock. Minerva Anestesiol. 2011;77:986–992. [PubMed] [Google Scholar]
- 40.Wheeler DS, Wong HR, Zingarelli B. Pediatric sepsis: part I. “Children are not small adults!”. Open Inflamm J. 2011;4:4–15. doi: 10.2174/1875041901104010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, Weiss SL, Fitzgerald J, Checchia PA, Meyer K, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med. 2015;191:309–315. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.del Fresno C, García-Rio F, Gómez-Piña V, Soares-Schanoski A, Fernández-Ruíz I, Jurado T, Kajiji T, Shu C, Marín E, Gutierrez del Arroyo A, et al. Potent phagocytic activity with impaired antigen presentation identifying lipopolysaccharide-tolerant human monocytes: demonstration in isolated monocytes from cystic fibrosis patients. J Immunol. 2009;182:6494–6507. doi: 10.4049/jimmunol.0803350. [DOI] [PubMed] [Google Scholar]
- 43.Pena OM, Pistolic J, Raj D, Fjell CD, Hancock RE. Endotoxin tolerance represents a distinctive state of alternative polarization (M2) in human mononuclear cells. J Immunol. 2011;186:7243–7254. doi: 10.4049/jimmunol.1001952. [DOI] [PubMed] [Google Scholar]
- 44.Conner SD, Schmid SL. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J Cell Biol. 2002;156:921–929. doi: 10.1083/jcb.200108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veiga E, Guttman JA, Bonazzi M, Boucrot E, Toledo-Arana A, Lin AE, Enninga J, Pizarro-Cerdá J, Finlay BB, Kirchhausen T, et al. Invasive and adherent bacterial pathogens co-opt host clathrin for infection. Cell Host Microbe. 2007;2:340–351. doi: 10.1016/j.chom.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Billingsley ML, Yun J, Reese BE, Davidson CE, Buck-Koehntop BA, Veglia G. Functional and structural properties of stannin: roles in cellular growth, selective toxicity, and mitochondrial responses to injury. J Cell Biochem. 2006;98:243–250. doi: 10.1002/jcb.20809. [DOI] [PubMed] [Google Scholar]