Advertisement
Research ArticleImmunology Free access | 10.1172/JCI44675
CD8+ T cells with an intraepithelial phenotype upregulate cytotoxic function upon influenza infection in human lung
Berber Piet,1,2 Godelieve J. de Bree,1 Barbara S. Smids-Dierdorp,1 Chris M. van der Loos,3 Ester B.M. Remmerswaal,1 Jan H. von der Thüsen,3 Jan M.W. van Haarst,4 Jan P. Eerenberg,5 Anja ten Brinke,6 Wim van der Bij,7 Wim Timens,8 René A.W. van Lier,1 and René E. Jonkers2
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by Piet, B. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by de Bree, G. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by Smids-Dierdorp, B. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by van der Loos, C. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by Remmerswaal, E. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by von der Thüsen, J. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by van Haarst, J. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by Eerenberg, J. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by ten Brinke, A. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by van der Bij, W. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by Timens, W. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by van Lier, R. in: JCI | PubMed | Google Scholar
1Department of Experimental Immunology, 2Department of Respiratory Medicine, and 3Department of Pathology, Academic Medical Centre, Amsterdam, Netherlands. 4Department of Respiratory Medicine and 5Department of Surgery, Tergooi Hospitals, Hilversum, Netherlands. 6Department of Immunopathology, Sanquin Research, Amsterdam, Netherlands. 7Department of Pulmonary Disease and Lung Transplantation and 8Department of Pathology and Medical Biology, University Medical Centre Groningen, Groningen, Netherlands.
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
Find articles by Jonkers, R. in: JCI | PubMed | Google Scholar
Published May 2, 2011 - More info
J Clin Invest. 2011;121(6):2254–2263. https://doi.org/10.1172/JCI44675.
© 2011 The American Society for Clinical Investigation
Received: August 9, 2010; Accepted: March 9, 2011
-
Abstract
The human lung T cell compartment contains many CD8+ T cells specific for respiratory viruses, suggesting that the lung is protected from recurring respiratory infections by a resident T cell pool. The entry site for respiratory viruses is the epithelium, in which a subset of lung CD8+ T cells expressing CD103 (αE integrin) resides. Here, we determined the specificity and function of CD103+CD8+ T cells in protecting human lung against viral infection. Mononuclear cells were isolated from human blood and lung resection samples. Variable numbers of CD103+CD8+ T cells were retrieved from the lung tissue. Interestingly, expression of CD103 was seen only in lung CD8+ T cells specific for influenza but not in those specific for EBV or CMV. CD103+ and influenza-reactive cells preferentially expressed NKG2A, an inhibitor of CD8+ T cell cytotoxic function. In contrast to CD103–CD8+ T cells, most CD103+CD8+ cells did not contain perforin or granzyme B. However, they could quickly upregulate these cytotoxic mediators when exposed to a type I IFN milieu or via contact with their specific antigen. This mechanism may provide a rapid and efficient response to influenza infection, without inducing cytotoxic damage to the delicate epithelial barrier.
-
Introduction
The lung parenchyma is only separated from the outside world by the 1-cell-layer thick epithelial barrier, which makes the lung a preferential entrance site for viruses. Respiratory viral infections are not only highly prevalent, they also form the main cause of virus-induced mortality in the Western world. The defence against viral infections is orchestrated by a cooperation of innate and adaptive immunity. One of the key components of the adaptive defence against viruses is the CD8+ T cell compartment. CD8+ T cells are abundantly present in both the airways and the lung parenchyma (1). They may kill virus-infected epithelial cells via the perforin/granzyme B pathway (cytotoxicity) or inhibit viral replication via secretion of IFN-γ (2–4). In mice it has been shown that the presence of local respiratory virus-specific memory T cells accelerates viral clearance and ameliorates survival upon secondary challenge with the same or related viruses (5–7). This is especially important for viruses like the influenza virus that can circumvent antibody-mediated immunity by mutation. Memory CD8+ T cells are mostly specific for highly conserved internal virus proteins and are therefore able to mount fast and efficient recall responses against a broad range of virus strains (8, 9). Previous research has shown that the human lung also contains pools of CD8+ T cells specific for respiratory viruses (10).
Human lung CD8+ T cells differ phenotypically from peripheral blood CD8+ T cells (10). Moreover, the local intraepithelial T cell pool (intraepithelial lymphocytes [IELs]) in the human lung is markedly different from subepithelial lamina propria T cells. Intraepithelial CD8+ T cells express αEβ7 integrin, which binds to epithelial cadherin (E-cadherin), expressed by the epithelial cells (11–16). This interaction retains the IELs in the vicinity of the epithelium (17–19). However, it is currently unknown whether the specificities and functions of lung CD8+ T cells differ between distinct anatomical locations, i.e., epithelium versus parenchyma. Due to practical and ethical limitations, most lung-related immunological research in humans is carried out on peripheral blood cells or on cells derived from the airway compartment (bronchoalveolar lavage fluid or sputum). In the present study, we analyzed human lung tissue-derived T cells by isolating them from fresh lung resection material. Our aim was to determine the specificity and function of CD103+CD8+ T cells. Our data suggest that the lung is equipped with a local virus-specific epithelial CD8+ T cell subset that might protect the lung against recurring influenza virus infection.
-
Results
Human lung CD8+ T cells expressing αE integrin are IELs. We collected paired peripheral blood and lung samples to be able to directly compare the expression of αE integrin (CD103) on lung CD8+ T cells with its expression on peripheral blood CD8+ T cells. We found a large and highly significant difference in CD103 expression on total CD3+CD8+ T cells between blood (mean of 1.9%) and lung (mean of 35%) (Figure 1, A and B).
 Figure 1
Figure 1CD103 expression on lung CD8+ T cells is highly increased compared with that on peripheral blood CD8+ T cells. (A) The percentages of peripheral blood CD8+ T cells and lung CD8+ T cells expressing CD103, as measured by flow cytometry, in paired peripheral blood and lung samples (n = 34). ***P < 0.0001; paired t test. (B) Representative histogram plots for the expression of CD103 on CD8+ T cells in the peripheral blood and human lung. Histogram plots show only CD3+CD8+ cells within the lymphocyte gate. The numbers in the plot represent the MFI of the CD103+ cells. (C) Preferential localization of CD103+CD8+ T cells above the basement membrane (dotted line) inside the airway epithelium (E). Original magnification, ×20. Immunohistochemistry stainings were performed on frozen human lung sections. To preserve the original tissue structure, we embedded the tissue in TissueTek prior to freezing. Representative images of airway containing tissue section. The image on the left is the original image; the image in the middle is a spectral analysis of all stained cells; and the image on the right is a spectral analysis of exclusively triple+ cells. Blue, CD3+; red, CD8+; green, CD103+; yellow, CD3/CD8/CD103 triple+ (middle and right images). n = 4 patients.
Immunohistochemistry confirmed that most lung CD103+CD3+CD8+ T cells were indeed located intraepithelially above the basement membrane of the small airways (Figure 1C), as was previously published (13–16).
In humans, the majority of IELs in the intestine is CD8αβ+, and only a minority is CD8γδ+ (18). To quantify the contribution of γδ T cells to the CD103+ lung intraepithelial T cell pool, we analyzed the expression of TCR-γδ on lung CD8+ T cells in a subgroup of patients. As shown in Supplemental Figure 1, around half of the γδ T cells expressed CD103, but overall, γδ T cells formed only a minor proportion (<5%) of both the CD103+ and CD103–CD8+ T cell fractions. Thus CD103+CD8+ T cells are mainly intraepithelial CD8αβ+ T lymphocytes and comprise around one-third of the total human lung CD8+ T cell population.
Influenza-specific but not CMV- or EBV-specific CD8+ T cells in the human lung have an intraepithelial phenotype. CD103+CD8+ T cells are located mainly inside the epithelium, which is the entrance site for respiratory viruses. Therefore, we tested whether these T cells could play a role specifically in the defence against respiratory viruses. Virus-specific cells were identified by HLA-peptide tetrameric complexes and enumerated by flow cytometry. The tetramers were loaded with peptides specific for the respiratory influenza virus (FLU) or the nonrespiratory EBV or CMV.
Strikingly, a considerable proportion of the FLU-specific lung CD8+ T cells did express CD103, in marked contrast to CMV- and EBV-specific lung CD8+ T cells, which lacked expression of the integrin (Figure 2, B and C, and Supplemental Figure 2A; supplemental material available online with this article; doi: 10.1172/JCI44675DS1). There was no difference in tetramer staining MFI between CD103+ and CD103–CD8+ T cells (data not shown). In addition, many FLU-specific cells in the lung expressed VLA-1 (α1β1 integrin) (Figure 2, B and C), which in mice has been shown to be responsible for retaining influenza-specific memory CD8+ T cells in the lung by binding to the type IV collagen-rich basement membrane (20–22). Although VLA-1 is expressed mainly by CD103+CD8+ lung T cells (15) (Supplemental Figure 3), both CD103+ and CD103– FLU-specific lung T cells expressed VLA-1. Thereby, the vast majority of FLU-specific T cells (mean of 87%) expressed at least 1 of these 2 integrins (Supplemental Figure 2B), whereas this was only the case for the minority of EBV- and CMV-specific T cells. Interestingly, a fraction of the FLU-specific T cells in the human lung expressed NKG2A (Figure 2, B and C), which is an inhibitory NK cell receptor that modulates CD8+ T cell effector function and that is known to be highly expressed by intraepithelial T cells in the intestine (23–28). CMV- and EBV-specific CD8+ T cells in the lung expressed only little VLA-1 and were NKG2A negative. These results indicate that lung CD8+ T cells specific for influenza virus are equipped with cell surface molecules that enable them to reside at the epithelial entrance site of this respiratory virus. This is in contrast with cells specific for the nonrespiratory viruses CMV and EBV, which strongly suggests that these cells are not located in the epithelium but are either residing deeper inside the lung parenchyma or are just passersby. Virus-specific T cells in the peripheral blood were CD103 negative, whether they were specific for FLU, EBV, or CMV (Figure 2C and Supplemental Figure 2). FLU-specific cells, but not EBV- and CMV-specific cells, in the periphery expressed NKG2A and VLA-1, albeit at lower levels than those expressed in the lung (Figure 2C and Supplemental Figure 2A).
 Figure 2
Figure 2Influenza-specific lung CD8+ T cells have an intraepithelial phenotype, expressing CD103, VLA-1, and NKG2A, whereas cells specific for nonrespiratory viruses lack the expression of these epithelial markers. (A) FACS plots showing tetramer stainings on lung CD3+ cells after gating on live cells. Numbers indicate the percentages of CD3+ cells that are tetramer+. (B) Percentages of influenza-specific, EBV-specific, and CMV-specific lung CD8+ T cells expressing CD103, NKG2A, and VLA-1, as assessed by flow cytometry (n = 5–9 per tetramer). Bars represent the median. (C) Representative FACS plots for phenotype of lung and peripheral blood CD8+ T cells. FACS plots show the expression of CD103, NKG2A, and VLA-1 on total lung CD8+ T cells (left column), on FLU- and EBV-specific lung CD8+ T cells (second and third columns, respectively), on total peripheral blood CD8+ T cells (fourth column), and on FLU- and EBV-specific peripheral blood CD8+ T cells (fifth and sixth columns, respectively). Numbers in first and fourth columns indicate the percentages of CD3+CD8+ cells that are CD103+, NKG2A+, or VLA-1+. Numbers in second, third, fifth, and sixth columns indicate the percentages of tetramer+ cells that are CD103+, NKG2A+, or VLA-1+. Plots are representative of 5 to 9 patients per tetramer (lung) or 2 to 6 patients per tetramer (peripheral blood). FLU-specific cells could often not be detected in the blood with tetramer staining, although these patients did have FLU+ cells in the lung. CD8+ T cells were all identified as CD3+CD8+ cells within the lymphocyte gate.
CD103+CD8+ T cells have an effector or memory phenotype and produce high amounts of IFN-γ and other Th1 cytokines. To correlate the possible cellular localization with CD8+ T cell phenotype and function, we first assessed the differentiation stage of the CD103+CD8+ T cells in the lung by performing FACS stainings for CD27 and CD45R0 (29). CD103+ and CD103–CD8+ T cells had either a memory (CD27+CD45R0+) or an effector (CD27–CD45R0±) phenotype (Figure 3A). A remarkable difference though is the absence of CD45R0– cells (naive cells and CD45R0– effector-type cells) within the CD103+ subset (Figure 3B).
 Figure 3
Figure 3Lung CD103+CD8+ T cells are CD45R0+ effector or memory cells. Differentiation stages of the CD8+ T cells within the CD103+ and CD103– cell fractions as assessed by flow cytometry. Naive cells were defined as CD27+CD45R0–, memory cells were defined as CD27+CD45R0+, and effector cells were defined as CD27–CD45R0±. (A) Representative FACS plots for CD103+ and CD103–CD8+ T cells. Dot plots are gated on CD3+CD8+CD103+ cells among live lymphocytes or on the CD3+CD8+CD103– cells among live lymphocytes. Numbers indicate the percentages of CD3+CD8+CD103+ or CD3+CD8+CD103– cells that are located within each quadrant. (B) Naive CD8+ T cells (CD27+CD45R0–), memory CD8+ T cells (CD27+CD45R0+), effector/memory (EM) CD8+ T cells (CD27–CD45R0±), and CD45R0–CD8+ T cells as percentages of total CD103+ and CD103–CD8+ T cell fractions (n = 28). ***P < 0.0001.
As lung CD8+ T cells are effector or memory cells, we examined their cytokine production by stimulating total lung mononuclear cells (LMCs) with phorbol 12-myristate 13-acetate (PMA) and ionomycin. This type of stimulation did not lead to upregulation of CD103 on CD8+ T cells (data not shown). The majority of lung CD8+ T cells produced IFN-γ, and a substantial portion of these cells also produced TNF-α and IL-2 (Figure 4A and Supplemental Figure 4). Interestingly, the percentages of cells producing IFN-γ or IL-2 were significantly higher in the CD103+CD8+ T cell subset than in the CD103–CD8+ T cell subset (Figure 4A). IL-17, IL-4, IL-10, and IL-22 were only produced by a very small number of cells (Figure 4A and Supplemental Figure 4). IL-22 was produced by a significantly greater fraction of CD103+CD8+ T cells, and IL-4 was produced by a larger fraction of CD103–CD8+ T cells. Peripheral blood CD8+ T cells just contained a very small population of CD103+ cytokine-producing CD8+ T cells (Supplemental Figure 5, A and C). Of the 3 main cytokines produced, only IL-2 was produced by more CD103+ than CD103–CD8+ peripheral blood T cells (Supplemental Figure 5B).
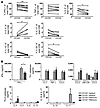 Figure 4
Figure 4CD103+ and CD103– lung CD8+ T cells produce high amounts of IFN-γ and other Th1 cytokines but hardly any Th2 cytokines, IL-10, or IL-17. CD103+CD8+ T cells produce more IFN-γ than the CD103–CD8+ T cell fraction. (A) Intracellular cytokine production of lung CD8+ T cells was measured by flow cytometry after 4-hour stimulation with PMA and ionomycin in the presence of brefeldin A. Cells were stained with life/dead stain (to gate out dead cells) and with monoclonal antibodies against CD3, CD8, and CD103 and fixed, permeabilized, and stained for intracellular cytokines. Graphs show the percentages of CD103+ and CD103– CD8+ T cells that produced each indicated cytokine. n = 5–10 patients. *P ≤ 0.04, **P = 0.003. (B) Cytokine production of CD103+ and CD103– lung CD8+ T cells as measured by luminex assay. CD103+ and CD103– lung CD8+ T cells from 4 patients were sorted with flow cytometry and stimulated for 24 hours with PMA and ionomycin (PMA/iono). Culture supernatants were collected and measured by the Bio-Rad 27-plex luminex assay. The error bars show SEM. *P = 0.03.
Second, purified lung CD103+ and CD103–CD8+ T cells were stimulated with PMA and ionomycin, and cytokine secretion was measured in the culture supernatants. Overall, CD103+ and CD103–CD8+ T cells showed a similar Th1 cytokine production profile, excreting IFN-γ, TNF-α, IL-2, CCL3 (MIP1α), CCL4 (MIP1β), CCL5 (RANTES), and GM-CSF (Figure 4B, top panels). Moreover, there was a significantly higher production of IFN-γ by the CD103+ cells, corroborating the data from the intracellular cytokine staining (Figure 4B). Various Th2 cytokines and IL-17 were hardly detectable, and IL-10 was not produced in relevant amounts by either cell fraction (Figure 4B, top panels). The latter finding is important, because previous research has shown that peripheral blood CD103+CD8+ T cells can have regulatory capacity by producing IL-10 (30). Overall, these data imply that both CD103+ and CD103–CD8+ T cells in the human lung have the ability to attract and activate many types of immune cells by the production of various Th1 type chemokines and cytokines. In addition, CD103+ cells appear to be more potent in producing IFN-γ.
CD103+CD8+ T cells have low expression of granzyme B and perforin. One of the main functions of CD8+ T cells is the exertion of cytotoxic activity to eliminate virus-infected cells. A prominent pathway for CD8-mediated cytotoxicity is the perforin/granzyme B axis (3, 4). After entering the target cell in a perforin-mediated way, granzyme B induces apoptosis of target cells (31, 32). Whereas CD103–CD8+ lung T cells expressed vast amounts of both perforin and granzyme B (mean expression 27% and 45%, respectively), their CD103+ counterparts hardly expressed any perforin and had a significantly reduced amount of granzyme B (mean expression 2.5% and 14%, respectively) (Figure 5, A and C). This difference was not caused by differences in the proportion of CD27– effector-type cells in the 2 CD8+ fractions, because these were comparable (Figure 3B). Peripheral blood CD103+CD8+ T cells also expressed less perforin and granzyme B than their CD103– counterparts (Supplemental Figure 6, A and B). However, this difference was less pronounced than that for lung CD8+ T cells and might be biased by the presence of recent thymic emigrants in the blood CD103+CD8+ T cell fraction (33).
 Figure 5
Figure 5CD8+CD103+ lung T cells have a low cytotoxic potential and a high expression of the inhibitory NK cell receptor complex CD94/NKG2A. (A) FACS plots show the expression of perforin, granzyme B, CD94, and NKG2A plotted against CD103 expression. Plots show only CD3+CD8+ lung T cells within the lymphocyte gate and are representative of 22–30 patients. Numbers indicate the percentages of CD3+CD8+ cells that are located within each quadrant. (B) Most lung CD8+ T cells express CD94 and NKG2A to the same extent, which is suggestive of dimerization (left). Numbers indicate the percentages of CD3+CD8+ cells that are located within each quadrant. Moreover, lung CD8+ T cells hardly express the activating isoform NKG2C, and there is no difference in NKG2C expression between CD103+ and CD103– T cells (right, n = 5). (C) The percentage of CD103+CD8+ and CD103–CD8+ lung T cells containing perforin or granzyme B (n = 22). (D) The percentage of CD103+CD8+ and CD103–CD8+ lung T cells expressing CD94 (n = 27) and NKG2A (n = 30). All expression data were collected with flow cytometry; all FACS plots show only lung CD3+CD8+ lymphocytes within the lymphocyte gate. ***P < 0.0001.
CD103+CD8+ T cells have high expression of the inhibitory NK cell receptor complex CD94/NKG2A. CD94/NKG2A is an inhibitory NK cell receptor complex that was first identified on NK cells but is also expressed on the surface of CD8+ T cells. Ligation of CD94/NKG2A with its nonclassical MHC class I ligand HLA-E leads to inhibition of cytotoxic degranulation and TCR-mediated TNF production (24–28). CD94/NKG2A has been shown to be highly expressed by IELs in the human intestine (23). In concordance with these gut-derived data, the expression of CD94 and NKG2A was significantly increased on the lung CD103+CD8+ T cell fraction compared with that on the lung CD103–CD8+ T cell fraction (Figure 5, A and D). Most lung CD8+ T cells expressed CD94 and NKG2A equally, which is suggestive of dimerization (Figure 5B, left panel). CD94 can also form a dimer with NKG2C, the activating isoform of NKG2A (34), but only a very small fraction of lung CD8+ T cells expressed NKG2C, and there was no difference in NKG2C expression between CD103+ and CD103– cells (Figure 5B, right panel). In peripheral blood, CD103+CD8+ T cells had a higher CD94 and NKG2A expression than CD103–CD8+ T cells (Supplemental Figure 6, A and B). Similar to the expression of granzyme B and perforin, this difference was much smaller than that in the lung. Overall, CD94+, NKG2A+, perforin–, and granzyme B– CD103+CD8+ T cells did not constitute a substantial population within the total peripheral blood CD8+ T cell pool (Supplemental Figure 6C). The high expression of CD94/NKG2A on CD103+ lung CD8+ T cells implies that these cells can be inhibited in their effector function when ligated to HLA-E. In addition to their low levels of perforin and granzyme B content, this might lead to a further reduction in their cytotoxic capacity.
TCR activation and type I IFNs induce upregulation of effector capacity in CD103+CD8+ T cells. To test whether the differences in the expression of cytotoxic mediators and inhibitory receptors between CD103+ and CD103–CD8+ T cells lead to a difference in cytotoxic function, we performed a redirected killing assay. We sorted pure cell populations of lung CD103+ and CD103–CD8+ T cells and determined cytotoxic T cell activity in an aCD3 mAb-mediated cytotoxicity assay. Surprisingly, CD103+CD8+ T cells proved to be even more potent in killing Fc-receptor bearing target cells than both CD103–CD8+ T cells and peripheral blood effector CD8+ T cells (Figure 6). This result implied that cytolytic effector molecules can be rapidly upregulated in CD103+ T cells after activation. Indeed, as early as 60 minutes after the start of aCD3 stimulation, intracellular FACS staining showed an upregulation of both perforin and granzyme B (data not shown). Thus, although CD103+CD8+ lung T cells do not express cytolytic effector molecules in vivo, they have the ability to quickly upregulate these mediators and execute cytolysis in vitro.
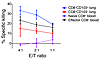 Figure 6
Figure 6CD8+CD103+ lung T cells have a high cytotoxic activity in vitro. The amount of specific killing of target cells by purified T cell populations at varying effector/target (E/T) ratios is shown. A redirected killing assay was performed by incubating FACS-sorted CD103+ or CD103– lung CD8+ T cells with chromium-labeled P815 target cells at varying effector/target ratios in the presence or absence of aCD3 mAb. FACS-sorted peripheral blood naive (CD45RA+CD27bright) and effector (CD27–) CD8+ T cells were included in this assay as control. Measurements were performed in triplicate (n = 3 patients). The mean ± SEM of specific killing is shown.
To test whether lung CD8+ T cells also upregulate granzyme B and perforin upon encounter with their specific antigen, we stimulated LMCs from donors who were known to react with the FLU-A2 tetramer for 6 days with FLU-A2 peptide. A mean of 90% of FLU+CD8+ T cells expressed perforin and a mean of 95% of FLU-A2+ CD8+ T cells expressed granzyme B after stimulation with FLU peptide (Figure 7A). This was significantly higher than the 30% and 24% of FLU-specific cells expressing perforin+ and granzyme B+, respectively, in the medium control or the 23% and 15% of FLU-specific cells expressing perforin+ and granzyme B+, respectively, (percentages are mean expression levels), after stimulation with an irrelevant peptide (Figure 7A). Recent research findings by Kohlmeier et al. showed that expression of granzyme B by virus-specific CD8+ T cells in mouse lungs and human peripheral blood could also be induced by antigen-independent stimulation with type I IFNs (35). To analyze whether, next to that of TCR/CD3 ligation, type I IFN could also induce granzyme B upregulation in human lung CD8+ T cells, we cultured human LMCs in the presence of polyinosinic-polycytidylic acid (poly I:C) (to induce endogenous type I IFN production) or varying concentrations of IFN-α. Strikingly, FLU-specific cells already showed a marked increase in granzyme B expression after 12 hours in any of these conditions (Figure 7B, left panel; mean increase in the 3 conditions compared with that of medium, control 61% to 154%), and this effect of IFN-α seemed to be dose dependent. Total lung CD8+ T cells also upregulated granzyme B protein expression in a type I IFN milieu, although this increase was less pronounced than that in the FLU-specific cells (Figure 7B, right panel). These data show that when influenza-specific lung CD8+ T cells are confronted with their specific antigen or an antigen-independent viral infectious milieu, they have the ability to rapidly upregulate cytotoxic proteins.
 Figure 7
Figure 7Lung CD8+ T cells quickly upregulate cytotoxic molecules after specific antigen contact or under influence of a type I IFN infectious milieu. (A) LMCs were cultured for 6 days in the presence of FLU peptide, irrelevant peptide (OVA peptide), or medium. FACS plots and bar graphs show the expression of perforin and granzyme B on FLU-A2+ cells (n = 2), as measured by flow cytometry. Plots are gated on CD3+CD8+ T cells and are representative of 2 patients. Numbers indicate the percentages of CD3+CD8+ cells located in each quadrant. (B) LMCs were stimulated for 12 or 36 hours with poly I:C or different concentrations of IFN-α. Granzyme B expression was measured by flow cytometry. The percentage increase in granzyme B expression as compared with that in medium control is shown for FLU+CD3+CD8+ T cells (left) and total CD3+CD8+ T cells (right) (n = 3 patients). The error bars show SEM. *P < 0.05, **P < 0.01.
-
Discussion
The respiratory influenza virus–specific CD8+ T cells in the human lung have an “intraepithelial fingerprint,” with high CD103, VLA-1, and NKG2A expression, which suggests that these cells are maintained preferentially close to the airway epithelium. This is in sharp contrast with CD8+ T cells in the lung specific for the nonrespiratory viruses CMV and EBV, which lack expression of CD103 and NKG2A, have a low expression of VLA-1, and are therefore more likely located in the parenchyma. We previously published that respiratory virus-specific T cells reside in the human lung, rather than in the peripheral blood (10), but their location within the lung had not been studied. The presence of virus-specific memory cells at the viral entrance site is important, since this can facilitate local antigen presentation, which could enable these virus-specific T cells to directly exert their effector function, without migration to the lymph node being necessary (36). This way the viral load and epithelial damage can be reduced before the recruitment of memory cells from the circulation and lymphoid tissue has taken place, thereby substantially reducing disease duration and pathology. In addition, immunopathology may be limited, as the initially reduced viral load will lead to a reduced degree of immune activation (37).
A limitation to our study is the fact that we analyzed the exact location of CD103+CD8+ lung T cells only in a subgroup of our patients. However, our findings in this group are comparable to data published by Hirosako et al., who have meticulously analyzed CD103+ expression by intraepithelial T cells in human bronchi (third to fifth generation) in a patient group that is demographically very similar to our cohort (14, 16). They showed that a range of 74% to 94% of intraepithelial CD8+ cells expressed CD103, whereas only 8% to 25% of lamina propria T cells expressed CD103. Although this means that there is not a one-on-one relationship between CD103 expression and intraepithelial location, the expression difference establishes CD103 as the best marker for distinguishing location-associated phenotypical and functional differences by flow cytometry.
A substantial fraction of the lung lymphocytes used in our study was derived from human lung tissue specimens obtained from patients undergoing a lobectomy for an isolated peripheral lung carcinoma. The peripheral lung tissue we used was sampled as far away as possible from the tumor. In these specimens, there were no macroscopic or microscopic abnormalities, and there were no signs of cellular infiltration. Furthermore, we found the same expression patterns of cytokines, CD103, CD94/NKG2A, VLA-1, granzyme B, and perforin on lung CD8+ T cells from the lung transplantation patients and our healthy donor, who were all free of malignancy. Therefore, we consider it highly unlikely that the phenotypical and functional characteristics of lung CD8+ T cells in our study were influenced by the presence of malignancy.
The environment plays an important role in maintaining local immunity. Several murine studies already showed that influenza-specific lung CD8+ T cells express VLA-1. Binding of VLA-1 to components of the ECM maintained virus-specific cells in nonlymphoid tissue by preventing migration and enhancing survival (20–22, 38, 39). The high VLA-1 expression that we find on both influenza-specific and intraepithelial CD8+ T cells in the human lung suggests evolutionary conservation of this mechanism and could well play a role in maintaining these cells in the vicinity of the port of entry of airway pathogens. The HLA-peptide tetramer technique we used for the identification of virus-specific CD8+ T cells will give an underestimation of the actual number of FLU-specific cells (40). Therefore, it is highly likely that influenza infection has a substantial impact on the formation of the epithelial CD8+ T cell pool.
A prominent finding of our study is the strong difference in the expression of cytotoxic mediators between CD103+ and CD103–CD8+ T cells. However, we also found that lung CD8+ T cells have the ability to rapidly upregulate perforin and granzyme B upon antigen-specific contact or in an antigen-independent viral infection (type I IFN) environment. In vitro, CD103+CD8+ T cells even surpass CD103– cells in cytotoxic killing capacity. CD103+ T cells have been shown before to be very potent cytotoxic killers in a number of other settings (41–46). Additionally, CD103+CD8+ lung T cells are also strong Th1 cytokine producers. However, as IFN-γ is a potent inhibitor of herpes virus but not influenza virus replication, T cells need their cytotoxic function to adequately combat infection with this respiratory virus (47). We hypothesize that in the absence of infection CD103+ T cells are inhibited in their cytotoxic effector function by the epithelial environment. Upon viral (re-)infection, specific and aspecific stimuli might quickly overcome this environmental inhibition, which ensures a fast and powerful cytotoxic T cell response.
The fact that FLU-specific cells, but not EBV- and CMV-specific cells, in the peripheral blood expressed NKG2A, albeit at lower levels than in the lung, suggests that not only organ but also virus specificity plays a role in modulating CD8+ T cell phenotype. NKG2A expression on peripheral blood CD8+ T cells does not solely reflect the localization specificity of cells, it also plays a role in regulating effector functions (23–28). Influenza is a virus that is usually cleared, whereas CMV and EBV are viruses that persist latently. Influenza-specific CD8+ T cells may upregulate NKG2A to inhibit cytotoxicity and prevent immunopathology after viral clearance, whereas EBV- and CMV-specific cells should maintain their cytotoxic capacity to combat the persisting virus (48). These data indicate that the phenotype of virus-specific T cells is determined both by organ-specific features and viral characteristics.
Increased knowledge of the lung CD103+CD8+ T cell compartment could lead to a better understanding of many diseases involving the respiratory epithelium, like COPD, asthma, airway infections, and bronchial carcinoma. For example, an increase in the activation threshold of epithelial CD8+ T cells could create favorable conditions for carcinogenesis at epithelial sites. Several studies have shown that diminished cytotoxic function and high CD94/NKG2A expression lead to a reduction in the killing of (pre-)malignant cells and therefore increase the risk for carcinogenesis (28, 49–51).
Apprehension of the epithelial localization of influenza-specific T cells contributes to the understanding of the first phase of the immune response upon secondary infection. The clinical importance of local respiratory virus-specific memory T cells for accelerated viral clearance and ameliorated survival upon secondary infection has extensively been shown (5–7). Memory CD8+ T cells retain the ability to mount recall responses against influenza strains after mutations (9). Therefore locally residing virus-specific T cells might be a good focus for the development of new therapies against the frequently mutating influenza viruses such as H1N1.
We showed that lung CD8+ T cells specific for influenza virus express CD103 and VLA-1, which suggests that they are mainly maintained inside the airway epithelium. This specific location would make them a very fast and efficient first line of defence upon reinfection. We also showed that CD103+CD8+ T cells form a distinct T cell subset, with a low expression of cytotoxic mediators and a high expression of the inhibitory NK cell receptor complex CD94/NKG2A. We suggest that this specific phenotype is created by the epithelial milieu to limit cytotoxicity-induced damage to the epithelial barrier. The ability of these cells to rapidly upregulate perforin and granzyme B upon virus infection rescues their cytotoxic effector function in situations in which lysis of virus-infected epithelial cells operates to limit viral spread.
-
Methods
Patients. Material from a total of 50 subjects (28 male and 22 female) was collected. The median age of subjects was 63 years (interquartile range, 54.8–72.3 years). Thirty-nine patients underwent a lobectomy for a peripheral primary lung tumor, ten received lung transplantation because of end-stage pulmonological disease, and one tissue specimen was obtained from a healthy lung donor (Table 1). Patients with a history of asthma or a recent (<4 weeks) lower respiratory tract infection were excluded from the study. Three of the COPD transplantation patients received low-dose (5–10 mg) prednisolone therapy, and the pulmonary fibrosis transplantation patient received high-dose (60 mg) prednisolone treatment. The other patients did not receive systemic corticosteroids or other systemic immunosuppressive therapy at the time of inclusion in the study or in the recent past. None of the patients recently received chemotherapy or radiotherapy. Two subjects were never-smokers; the others were all (ex)smokers, of whom 18 still had smoked in the last 6 months prior to the moment we obtained the blood and lung specimens. The demographics of our cohort are comparable to those of cohorts used for previously published human lung CD8+ T cell studies (52). Lobectomy patients were recruited from the Academic Medical Centre and the Tergooi Hospitals. Lung transplantation patients were recruited from the University Medical Centre Groningen. All patients gave written informed consent prior to inclusion in the study, and the study was approved by the Ethical Review Board (ERB) of the Academic Medical Centre and the local ERBs of the other participating centres.
Isolation of mononuclear cells from peripheral blood and lung tissue. Heparinized peripheral blood samples were obtained prior to or during the surgical procedure. PBMCs were isolated using standard density gradient techniques. Directly after lobectomy, a piece of peripheral lung tissue, as far away from the tumor as possible, was cut off by a pathologist. LMCs were isolated from this tissue specimen, as described by Holt et al. (53). In brief, tissue specimens (1 × 1 cm) were sliced with a McIlwain tissue chopper into 1-mm pieces and incubated for 20 minutes in RPMI with 20 mM HEPES, 15% FCS, 50 U/ml DNAse type I (Sigma-Aldrich) while shaking at 37°C. Tissue pieces were carefully dried with sterile gauzes and transferred to medium supplemented with collagenase type I 300 U/ml (Worthington). The material was incubated in this medium for 60 minutes while shaking at 37°C. A cell suspension was obtained by grinding the tissue through a flow-through chamber. Mononuclear cells were isolated from the lung cell suspension by standard density gradient techniques. To exclude the possibility of contamination with peripheral blood, the erythrocyte counts were confirmed to be less than 5% of erythrocyte counts in the paired blood sample. Standard extra- and intracellular stainings were performed on freshly isolated LMCs, while tetramer stainings and stimulation experiments were performed on cells that were cryopreserved in liquid nitrogen for later analysis.
Immunohistochemistry. Small sections of the fresh lung material were embedded in TissueTek for optimal preservation of the original tissue structure and were subsequently snap frozen by short exposition to liquid nitrogen. Five-μm cryostat lung tissue sections were acetone-fixed and air dried. The sequential triple immunohistological staining started with aCD8 (rabbit IgG, clone SP16, Thermo Fischer Scientific) and aCD103 (mouse IgG1, clone Ber-ACT8, Dako), followed by anti-rabbit IgG/alkaline phosphatase–labeled (AP-labeled) polymer and anti-mouse IgG/HRP-labeled polymer (both from ImmunoLogic). AP activity was demonstrated with Vector Red (Vector Laboratories), and HRP activity was demonstrated by brown staining with Bright DAB (ImmunoLogic). The slides were shortly fixed with neutral buffered formalin and subjected to a heat-induced antigen retrieval procedure in citrate pH 6.0. This heat step removes and inactivates immunoreagents used in the first staining sequence, preventing unwanted cross-reactivity with the second immunohistochemistry staining sequence. Next, the slides were incubated with aCD3 monoclonal antibody (rabbit IgG, clone SP7, Thermo Fisher Scientific), followed by anti-rabbit IgG/AP-labeled polymer (ImmunoLogic). AP activity was demonstrated with Vector Blue (Vector Laboratories). Slides were fully dried with a hot plate and coverslipped with VectaMount (Vector Laboratories). The red, brown, and blue chromogens in the tissue sections were unmixed by the Nuance spectral imaging system (Cambridge Research Instrumentation) and imaged, according to the method for fluorescent images, with the Nuance 2.10 software in pseudocolors (54, 55).
Tetrameric complexes. The following APC-conjugated HLA-peptide tetrameric complexes were used: HLA-A1 tetramer loaded with FLU NP-derived CTELKLSDY peptide or CMV pp65-derived YSEHPTFTSQY peptide; HLA-A2 tetramer loaded with FLU M1-derived GILGFVFTL peptide, EBV BMLF1-derived GLCTLVAML peptide, or CMV IE1-derived VLEETSVML peptide; HLA-A11 tetramer loaded with EBV EBNA4-derived IVTDFSVIK peptide; HLA-B7 tetramer loaded with EBV EBNA3A-derived RPPIFIRRL peptide or CMV pp65-derived TPRVTGGGAM peptide; HLA-B8 tetramer loaded with EBV EBNA3A-derived FLRGRAYGL peptide, EBV BZLF1-derived RAKFKQLL peptide, CMV IE1-derived ELKRKMIYM peptide, CMV IE1-derived ELRRKMMYM peptide, or CMV IE1-derived QIKVRVDMV peptide; and HLA-B35 tetramer loaded with EBV EBNA1-derived HPVGEADYFEY peptide or CMV pp65-derived IPSINVHHY peptide. All tetramers were provided by Sanquin.
Flow cytometric analysis. PBMCs or LMCs (0.3 × 106 to 1.0 × 106 cells) were incubated with tetrameric complexes and different combinations of the following antibodies: aCD103 FITC (BD), aTCRγδ FITC (Immunotech), aCD103 PE (eBioscience), aCD27 PE (BD), aNKG2A PE (Beckman), aNKG2C PE (R&D Systems), aCD49a PE (BD Pharmingen), aCD45RA PerCP-Cy 5.5 (eBioscience), aCD8 PerCP-Cy 5.5 (BD), aCD8 PE–Alexa Fluor 610 (Invitrogen), aCD3 PE-Cy 7 (BD), aCD45R0 APC (BD Pharmingen), aCD94 APC (BD Pharmingen), aCD27 APC–Alexa Fluor 750 (eBioscience), and aCD27 APC-eFluor 780 (eBioscience). Cells were labeled according to the manufacturers’ instructions and washed and analyzed in PBS containing 0.01% (w/v) NaN3 and 0.5% (w/v) bovine serum albumin (PBA). For intracellular perforin, granzyme B, and cytokine staining, the following technique was used: after extracellular staining on 0.5 × 106 PBMCs or LMCs, cells were washed, fixed with 2% PFA solution, and permeabilized by washing with PBA with 0.1% saponin. Subsequently, cells were incubated with 1 or more of the following antibodies: anti-perforin FITC (BD), aTNF-α FITC (BD), anti-granzyme B PE (Sanquin), aIL-17A PE (eBioscience), aIL-22 PE (R&D Systems), aIFN-γ PerCP-Cy5.5 (eBioscience), aIL-2 APC (BD), aIL-4 APC (BD Pharmingen), and aIL-10 APC (BD Pharmingen). Cells were analyzed by FACSCanto (BD Biosciences) multicolor flow cytometry and FlowJo software (Tree Star Inc.).
Stimulation of LMCs and PBMCs. For intracellular cytokine staining, cells were stimulated at 37°C in culture medium (RPMI with 10% [w/v] HPS and antibiotics) for 4 hours with PMA (2 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich) in the presence of brefeldin A (10 μg/ml; Invitrogen).
For FLU peptide stimulations, LMCs were stimulated at 37°C in culture medium (RPMI with 10% [w/v] HPS and antibiotics) for 6 days with IL-2 (50 U/ml; Biotest Ag) and FLU-A2 peptide (1.25 μg/ml; Microbix Biosystems Inc.). As control, we cultured LMCs for 6 days at 37°C in culture medium with either IL-2 alone or IL-2 and an irrelevant peptide (OVA).
Stimulations with IFN-α (IFNα2a, PBL InterferonSource) and poly I:C (Sigma-Aldrich) were performed by stimulating LMCs for 12 or 36 hours with recombinant human IFN-α in an end concentration of 5,000 or 20,000 U/ml or poly I:C in an end concentration of 20 μg/ml.
Luminex assay. To obtain pure cell populations of either CD103+CD8+ T cells or CD103–CD8+ T cells, cells were sorted using a FACSAria (BD Biosciences). Sorted cells were stimulated at 37°C in culture medium (RPMI with 10% [w/v] HPS and antibiotics) for 24 hours with PMA (2 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich). Supernatants were collected and analyzed by Bioplex human cytokine 27-plex luminex assay (Bio-Rad) according to manufacturer’s instructions. This assay was also performed on the supernatants of the FLU-A2 peptide stimulations.
Redirected killing assay. Cytotoxic T cell activity was determined in an aCD3 mAb-mediated cytotoxicity assay as previously described (56). In brief, FcR-bearing P815 target cells were radiolabeled with Na51CrO4 (PerkinElmer) for 30 minutes at 37°C. Purified CD8+ T cell subsets were incubated with P815 target cells at varying effector/target ratios in the presence or absence of 5 μg/ml aCD3 mAb (CLB-CD3 1X1). After a 4-hour incubation period at 37°C, the supernatants of triplicate cultures were collected and counted in a gamma counter. Specific cytotoxicity was determined according to the following formula: percentage of specific lysis = 100 × ([cpm experimental release – cpm spontaneous release]/[cpm maximal release – cpm spontaneous release]). To obtain pure cell populations of lung CD103+CD8+ T cells, lung CD103–CD8+ T cells and peripheral blood naive (CD45RA+CD27bright) and effector (CD27–) CD8+ T cells, cells were sorted using a FACSAria (BD Biosciences).
Statistics. Paired samples that showed a Gaussian distribution were analyzed by the paired t test, and paired samples without Gaussian distribution were analyzed with the Wilcoxon signed-rank test. Unpaired samples were analyzed with the unpaired t test or the Mann-Whitney test. All t tests were 2 tailed. P < 0.05 was considered statistically significant.
-
Acknowledgments
We would like to thank Martijn Nolte and Kris Reedquist for critically reading the manuscript, Ineke ten Berge for helpful discussions, Berend Hooibrink for technical assistance with the FACS sorting, and Gerrit-Jan Ilbrink for asking for informed consent. We would furthermore like to express our gratitude to Jaap Kloek, Marnix Jonker, Eddy Hendriks, Jan Weening, Herbert Stel, Eric Prins, George Nossent, and Erik Verschuuren for help with obtaining patient material and to René Lutter for coordinating technical assistance. To conclude, we would like to thank the Netherlands Asthma Foundation for funding this research project (grant no. 3.2.06.020).
Address correspondence to: B. Piet, Department of Experimental Immunology, Meibergdreef 9/K0-152, 1105 AZ, Amsterdam, Netherlands. Phone: 31.20.5668041; Fax: 31.20.5669756; E-mail: b.piet@amc.nl.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2011;121(6):2254–2263. doi:10.1172/JCI44675.
Godelieve J. de Bree’s present address is: Department of Internal Medicine, Academic Medical Centre, Amsterdam, Netherlands.
Jan H. von der Thüsen’s present address is: Department of Histopathology, Royal Brompton and Harefield NHS Trust, London, United Kingdom.
René A.W. van Lier’s present address is: Division of Research, Sanquin Blood Supply, Amsterdam, Netherlands.
-
References
- Hogg JC, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(26):2645–2653.
- Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol. 1997;159(11):5197–5200.View this article via: PubMed Google Scholar
- Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol. 2002;20:323–370.
- Trapani JA, Sutton VR, Smyth MJ. CTL granules: evolution of vesicles essential for combating virus infections. Immunol Today. 1999;20(8):351–356.
- Cerwenka A, Morgan TM, Dutton RW. Naive, effector, and memory CD8 T cells in protection against pulmonary influenza virus infection: homing properties rather than initial frequencies are crucial. J Immunol. 1999;163(10):5535–5543.View this article via: PubMed Google Scholar
- Hogan RJ, Zhong W, Usherwood EJ, Cookenham T, Roberts AD, Woodland DL. Protection from respiratory virus infections can be mediated by antigen-specific CD4(+) T cells that persist in the lungs. J Exp Med. 2001;193(8):981–986.
- Liang S, Mozdzanowska K, Palladino G, Gerhard W. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J Immunol. 1994;152(4):1653–1661.View this article via: PubMed Google Scholar
- Ely KH, Roberts AD, Woodland DL. Cutting edge: effector memory CD8+ T cells in the lung airways retain the potential to mediate recall responses. J Immunol. 2003;171(7):3338–3342.View this article via: PubMed Google Scholar
- Kohlmeier JE, Woodland DL. Immunity to respiratory viruses. Annu Rev Immunol. 2009;27:61–82.
- de Bree GJ, van Leeuwen EM, Out TA, Jansen HM, Jonkers RE, van Lier RA. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J Exp Med. 2005;202(10):1433–1442.View this article via: PubMed Google Scholar
- Cepek KL, et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature. 1994;372(6502):190–193.
- Cerf-Bensussan N, Begue B, Gagnon J, Meo T. The human intraepithelial lymphocyte marker HML-1 is an integrin consisting of a beta 7 subunit associated with a distinctive alpha chain. Eur J Immunol. 1992;22(1):273–277.View this article via: PubMed Google Scholar
- Goto E, et al. Human bronchial intraepithelial T lymphocytes as a distinct T-cell subset: their long-term survival in SCID-Hu chimeras. Am J Respir Cell Mol Biol. 2000;22(4):405–411.View this article via: PubMed Google Scholar
- Hirosako S, et al. Human bronchial intraepithelial T cells produce interferon-gamma and stimulate epithelial cells. Clin Exp Immunol. 2009;155(2):266–274.
- Morgan AJ, Guillen C, Symon FA, Birring SS, Campbell JJ, Wardlaw AJ. CXCR6 identifies a putative population of retained human lung T cells characterised by co-expression of activation markers. Immunobiology. 2008;213(7):599–608.
- Hirosako S, et al. CD8 and CD103 Are highly expressed in asthmatic bronchial intraepithelial lymphocytes. Int Arch Allergy Immunol. 2010;153(2):157–165.
- El-Asady R, et al. TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med. 2005;201(10):1647–1657.
- Johansson-Lindbom B, Agace WW. Generation of gut-homing T cells and their localization to the small intestinal mucosa. Immunol Rev. 2007;215:226–242.
- Lefrancois L, et al. The role of beta7 integrins in CD8 T cell trafficking during an antiviral immune response. J Exp Med. 1999;189(10):1631–1638.
- Ray SJ, et al. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity. 2004;20(2):167–179.
- Richter M, et al. Collagen distribution and expression of collagen-binding alpha1beta1 (VLA-1) and alpha2beta1 (VLA-2) integrins on CD4 and CD8 T cells during influenza infection. J Immunol. 2007;178(7):4506–4516.View this article via: PubMed Google Scholar
- Chapman TJ, Topham DJ. Identification of a unique population of tissue-memory CD4+ T cells in the airways after influenza infection that is dependent on the integrin VLA-1. J Immunol. 2010;184(7):3841–3849.View this article via: PubMed Google Scholar
- Jabri B, et al. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology. 2000;118(5):867–879.
- Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795–799.View this article via: PubMed Google Scholar
- Figueiredo C, Seltsam A, Blasczyk R. Permanent silencing of NKG2A expression for cell-based therapeutics. J Mol Med. 2009;87(2):199–210.
- Ikeda H, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6(2):199–208.
- Mingari MC, Ponte M, Vitale C, Bellomo R, Moretta L. Expression of HLA class I-specific inhibitory receptors in human cytolytic T lymphocytes: a regulated mechanism that controls T-cell activation and function. Hum Immunol. 2000;61(1):44–50.
- Speiser DE, et al. In vivo expression of natural killer cell inhibitory receptors by human melanoma-specific cytolytic T lymphocytes. J Exp Med. 1999;190(6):775–782.View this article via: PubMed Google Scholar
- Hamann D, et al. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med. 1997;186(9):1407–1418.
- Uss E, Rowshani AT, Hooibrink B, Lardy NM, van Lier RA, ten Berge IJ. CD103 is a marker for alloantigen-induced regulatory CD8+ T cells. J Immunol. 2006;177(5):2775–2783.View this article via: PubMed Google Scholar
- Smyth MJ, Trapani JA. Granzymes: exogenous proteinases that induce target cell apoptosis. Immunol Today. 1995;16(4):202–206.
- Trapani JA, Davis J, Sutton VR, Smyth MJ. Proapoptotic functions of cytotoxic lymphocyte granule constituents in vitro and in vivo. Curr Opin Immunol. 2000;12(3):323–329.
- McFarland RD, Douek DC, Koup RA, Picker LJ. Identification of a human recent thymic emigrant phenotype. Proc Natl Acad Sci U S A. 2000;97(8):4215–4220.
- Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8(6):693–701.
- Kohlmeier JE, Cookenham T, Roberts AD, Miller SC, Woodland DL. Type I interferons regulate cytolytic activity of memory CD8+ T cells in the lung airways during respiratory virus challenge. Immunity. 2010;33(1):96–105.
- Wakim LM, Waithman J, van Rooijen N, Heath WR, Carbone FR. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 2008;319(5860):198–202.View this article via: PubMed Google Scholar
- Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol. 2009;9(3):153–161.
- Richter MV, Topham DJ. The alpha1beta1 integrin and TNF receptor II protect airway CD8+ effector T cells from apoptosis during influenza infection. J Immunol. 2007;179(8):5054–5063.View this article via: PubMed Google Scholar
- Wang XZ, Stepp SE, Brehm MA, Chen HD, Selin LK, Welsh RM. Virus-specific CD8 T cells in peripheral tissues are more resistant to apoptosis than those in lymphoid organs. Immunity. 2003;18(5):631–642.
- Lawrence CW, Ream RM, Braciale TJ. Frequency, specificity, and sites of expansion of CD8(+) T cells during primary influenza virus infection. J Immunol. 2005;174(9):5332–5340.View this article via: PubMed Google Scholar
- Franciszkiewicz K, et al. Intratumoral induction of CD103 triggers tumor-specific CTL function and CCR5-dependent T-cell retention. Cancer Res. 2009;69(15):6249–6255.
- French JJ, Cresswell J, Wong WK, Seymour K, Charnley RM, Kirby JA. T cell adhesion and cytolysis of pancreatic cancer cells: a role for E-cadherin in immunotherapy? Br J Cancer. 2002;87(9):1034–1041.
- Le Floc’h A, et al. Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. J Exp Med. 2007;204(3):559–570.
- Smyth LJ, Kirby JA, Cunningham AC. Role of the mucosal integrin alpha(E)(CD103)beta(7) in tissue-restricted cytotoxicity. Clin Exp Immunol. 2007;149(1):162–170.View this article via: PubMed Google Scholar
- Hadley GA, Charandee C, Weir MR, Wang D, Bartlett ST, Drachenberg CB. CD103+ CTL accumulate within the graft epithelium during clinical renal allograft rejection. Transplantation. 2001;72(9):1548–1555.
- Taunk J, Roberts AI, Ebert EC. Spontaneous cytotoxicity of human intraepithelial lymphocytes against epithelial cell tumors. Gastroenterology. 1992;102(1):69–75.View this article via: PubMed Google Scholar
- La Gruta NL, Kedzierska K, Stambas J, Doherty PC. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol. 2007;85(2):85–92.
- Zhou J, Matsuoka M, Cantor H, Homer R, Enelow RI. Cutting edge: engagement of NKG2A on CD8+ effector T cells limits immunopathology in influenza pneumonia. J Immunol. 2008;180(1):25–29.View this article via: PubMed Google Scholar
- Epling-Burnette PK, et al. Dysregulated NK receptor expression in patients with lymphoproliferative disease of granular lymphocytes. Blood. 2004;103(9):3431–3439.View this article via: PubMed Google Scholar
- Sheu BC, et al. Up-regulation of inhibitory natural killer receptors CD94/NKG2A with suppressed intracellular perforin expression of tumor-infiltrating CD8+ T lymphocytes in human cervical carcinoma. Cancer Res. 2005;65(7):2921–2929.
- Welsh RM, Stepp SE, Szomolanyi-Tsuda E, Peacock CD. Tumor viral escape from inhibited T cells. Nat Immunol. 2002;3(2):112–114.
- Freeman CM, et al. Cytotoxic potential of lung CD8(+) T cells increases with chronic obstructive pulmonary disease severity and with in vitro stimulation by IL-18 or IL-15. J Immunol. 2010;184(11):6504–6513.
- Holt PG, et al. Extraction of immune and inflammatory cells from human lung parenchyma: evaluation of an enzymatic digestion procedure. Clin Exp Immunol. 1986;66(1):188–200.View this article via: PubMed Google Scholar
- van der Loos CM. Multiple immunoenzyme staining: methods and visualizations for the observation with spectral imaging. J Histochem Cytochem. 2008;56(4):313–328.View this article via: PubMed Google Scholar
- van der Loos CM. Chromogens in multiple immunohistochemical staining used for visual assessment and spectral imaging: the colourful future. J Histotechnol. 2010;33(1):31–40.
- de Jong R, Brouwer M, Rebel VI, van Seventer GA, Miedema F, van Lier RA. Generation of alloreactive cytolytic T lymphocytes by immobilized anti-CD3 monoclonal antibodies. Analysis of requirements for human cytolytic T-lymphocyte differentiation. Immunology. 1990;70(3):357–364.View this article via: PubMed Google Scholar
-
Version history
- Version 1 (May 2, 2011): No description
- Version 2 (June 1, 2011): No description



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)

