MARK3
Appearance
| MARK3 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MARK3, CTAK1, KP78, PAR1A, Par-1a, microtubule affinity regulating kinase 3, VIPB | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 602678; MGI: 1341865; HomoloGene: 55653; GeneCards: MARK3; OMA:MARK3 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
MAP/microtubule affinity-regulating kinase 3 is an enzyme that in humans is encoded by the MARK3 gene.[5][6]
Interactions
[edit]MARK3 has been shown to interact with Stratifin.[7][8]
It has been linked to a form of genetic blindness, believed to be a genetic recessive disease that progressively destroys the eyes.[9]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000075413 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000007411 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
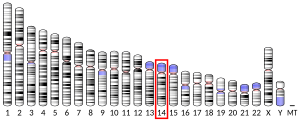
- ^ Ono T, Kawabe T, Sonta S, Okamoto T (April 1998). "Assignment of MARK3 alias KP78 to human chromosome band 14q32.3 by in situ hybridization". Cytogenet Cell Genet. 79 (1–2): 101–2. doi:10.1159/000134692. PMID 9533022.
- ^ "Entrez Gene: MARK3 MAP/microtubule affinity-regulating kinase 3".
- ^ Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein–protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- ^ Benzinger A, Muster N, Koch HB, Yates JR, Hermeking H (June 2005). "Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer". Mol. Cell. Proteomics. 4 (6): 785–95. doi:10.1074/mcp.M500021-MCP200. PMID 15778465.
- ^ "Découverte d'un gène provoquant une cécité - Communiqués de presse - UNIGE". 23 July 2018.
Further reading
[edit]- Tassan JP, Le Goff X (2004). "An overview of the KIN1/PAR-1/MARK kinase family". Biol. Cell. 96 (3): 193–9. doi:10.1016/j.biolcel.2003.10.009. PMID 15182702.
- Meller N, Liu YC, Collins TL, Bonnefoy-Bérard N, Baier G, Isakov N, Altman A (1996). "Direct interaction between protein kinase C theta (PKC theta) and 14-3-3 tau in T cells: 14-3-3 overexpression results in inhibition of PKC theta translocation and function". Mol. Cell. Biol. 16 (10): 5782–91. doi:10.1128/MCB.16.10.5782. PMC 231579. PMID 8816492.
- Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ, Wu Z, Stephenson MT, Piwnica-Worms H (1998). "C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding". Cell Growth Differ. 9 (3): 197–208. PMID 9543386.
- Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT (2001). "PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling". Nat. Cell Biol. 3 (7): 628–36. doi:10.1038/35083016. PMID 11433294. S2CID 1128604.
- Spicer J, Rayter S, Young N, Elliott R, Ashworth A, Smith D (2003). "Regulation of the Wnt signalling component PAR1A by the Peutz-Jeghers syndrome kinase LKB1". Oncogene. 22 (30): 4752–6. doi:10.1038/sj.onc.1206669. PMID 12879020. S2CID 9945293.
- Müller J, Ritt DA, Copeland TD, Morrison DK (2003). "Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate". EMBO J. 22 (17): 4431–42. doi:10.1093/emboj/cdg426. PMC 202368. PMID 12941695.
- Trinczek B, Brajenovic M, Ebneth A, Drewes G (2004). "MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes". J. Biol. Chem. 279 (7): 5915–23. doi:10.1074/jbc.M304528200. PMID 14594945.
- Navarro-Lérida I, Martínez Moreno M, Roncal F, Gavilanes F, Albar JP, Rodríguez-Crespo I (2004). "Proteomic identification of brain proteins that interact with dynein light chain LC8". Proteomics. 4 (2): 339–46. doi:10.1002/pmic.200300528. PMID 14760703. S2CID 8868600.
- Bachmann M, Hennemann H, Xing PX, Hoffmann I, Möröy T (2005). "The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint". J. Biol. Chem. 279 (46): 48319–28. doi:10.1074/jbc.M404440200. PMID 15319445.
- Benzinger A, Muster N, Koch HB, Yates JR, Hermeking H (2005). "Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer". Mol. Cell. Proteomics. 4 (6): 785–95. doi:10.1074/mcp.M500021-MCP200. PMID 15778465.
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE (2005). "A human protein–protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. hdl:11858/00-001M-0000-0010-8592-0. PMID 16169070. S2CID 8235923.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (2005). "Towards a proteome-scale map of the human protein–protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Göransson O, Deak M, Wullschleger S, Morrice NA, Prescott AR, Alessi DR (2007). "Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation". J. Cell Sci. 119 (Pt 19): 4059–70. doi:10.1242/jcs.03097. PMID 16968750. S2CID 582486.
- Dequiedt F, Martin M, Von Blume J, Vertommen D, Lecomte E, Mari N, Heinen MF, Bachmann M, Twizere JC, Huang MC, Rider MH, Piwnica-Worms H, Seufferlein T, Kettmann R (2006). "New role for hPar-1 kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases" (PDF). Mol. Cell. Biol. 26 (19): 7086–102. doi:10.1128/MCB.00231-06. PMC 1592903. PMID 16980613.
- Wissing J, Jänsch L, Nimtz M, Dieterich G, Hornberger R, Kéri G, Wehland J, Daub H (2007). "Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry". Mol. Cell. Proteomics. 6 (3): 537–47. doi:10.1074/mcp.T600062-MCP200. hdl:10033/19756. PMID 17192257.
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D (2007). "Large-scale mapping of human protein–protein interactions by mass spectrometry". Mol. Syst. Biol. 3 (1): 89. doi:10.1038/msb4100134. PMC 1847948. PMID 17353931.