Abstract
Mitogen-activated protein (MAP) kinases are central components of signal transduction pathways for cell proliferation, stress responses, and differentiation. Signaling efficiency and specificity are modulated in large part by docking interactions between individual MAP kinase and the kinase interaction motif (KIM), (R/K)2–3-X1–6-ΦA-X-ΦB, in its cognate kinases, phosphatases, scaffolding proteins, and substrates. We have determined the crystal structure of extracellular signal-regulated protein kinase 2 bound to the KIM peptide from MAP kinase phosphatase 3, an extracellular signal-regulated protein kinase 2-specific phosphatase. The structure reveals that the KIM docking site, situated in a noncatalytic region opposite of the kinase catalytic pocket, is comprised of a highly acidic patch and a hydrophobic groove, which engage the basic and ΦA-X-ΦB residues, respectively, in the KIM sequence. The specific docking interactions observed in the structure consolidate all known biochemical data. In addition, structural comparison indicates that the KIM docking site is conserved in all MAP kinases. The results establish a structural model for understanding how MAP kinases interact with their regulators and substrates and provide new insights into how MAP kinase docking specificity can be achieved.
Keywords: mitogen-activated protein kinase specificity
Mitogen-activated protein (MAP) kinase pathways serve as transducers of extracellular signals to enable cellular adaptation to changes in the surrounding environment (1, 2). These pathways are evolutionarily conserved and play essential roles in diverse physiological processes such as growth, differentiation, inflammatory responses, and programmed cell death. The three best characterized MAP kinase cascades are the extracellular signal-regulated protein kinase (ERK) pathway, which responds to stimuli that induce cell proliferation and differentiation, the c-Jun N-terminal protein kinase (JNK) pathway, and the p38 kinase pathway, both of which are activated in response to environmental stresses.
Given the importance of MAP kinase in cellular signaling, the activity and specificity of the MAP kinase must be tightly regulated to ensure proper integration of diverse biological stimuli and generation of appropriate cellular responses. The significant structural similarity among the multiple MAP kinase cascades in a cell raises an important question: how is signaling specificity achieved in the MAP kinase-mediated processes? Recent studies suggest that one mechanism to confer specificity and efficiency in MAP kinase signaling is through docking interactions between individual MAP kinases and their cognate activating kinases, inactivating phosphatases, scaffolding proteins, and substrates (3–15). A kinase interaction motif (KIM) or docking sequence (D domain), (R/K)2–3-X1–6-ΦA-X-ΦB, has been recognized in MAP kinase/ERK kinases, MAP kinase phosphatases (MKPs), scaffolding proteins, and MAP kinase substrates, where ΦA and ΦB are hydrophobic residues such as Leu, Ile, or Val, and X is any amino acid (Table 1). A common docking domain in MAP kinases composed of a stretch of negatively charged amino acids (e.g., Asp-316 and Asp-319 in ERK2) has been identified that may interact with the positively charged residues in KIM through electrostatic interactions (12, 14, 16, 17).
Table 1.
Sequence alignments of the KIM sequences from MKPs, kinases, scaffolding protein, and substrates
| Docking protein | KIM sequence | MAP kinase |
|---|---|---|
| MKP3 | PGIMLRRLQKGNLPVR | ERK |
| MKPX | PGLMLRRLRKGNLPIR | ERK |
| MKP4 | PALLLRRLRRGSLSVR | ERK |
| HVH5 | SKLVKRRLQQGKVTI | p38, JNK |
| MKP5 | CADKISRRRLQQGKITV | p38, JNK |
| MKP2 | RCNTIVRRRA–KGSVSLE | ERK, p38, JNK |
| EC-PTP | GLQERRGSNVSLTLDM | ERK |
| He-PTP | RLQERRGSNVALMLDV | ERK |
| STEP | GLQERRGSNVSLTLDM | ERK |
| JIP1 | RPKRP––TTLNLF | JNK |
| MEK1 | MPKKKP––TPIQLNPNP | ERK |
| MKK3b | KGSKRKK––DLRLSCNS | p38 |
| SEK1/MKK4 | QGKRKA–––LKLNF | JNK, p38 |
| MEF2A | RKPDLR–VVIPPS | p38 |
| RSK1 | SSILAQRRVRK––LPSTTL | ERK |
| MNK2 | QSKLAQRRQRAS–LSATPV | ERK |
| MNK1 | KSRLARRRALAQAGRSRD | ERK, p38 |
| MSK1 | KAPLAKRRKMKKTSTSTE | ERK, p38 |
| MAPKAPK2 | NPLLLKRRKKARALEAAA | p38 |
Basic residues and ΦA and ΦB are in bold.
There is intense interest in understanding the structural basis for docking interactions between MAP kinases and their cognate binding proteins. Unfortunately, no three-dimensional structure is available for any MAP kinase in complex with its effector molecules. However, the crystal structures of p38α bound with the KIM sequences derived from its activating kinase, MKK3b, and substrate, MEF2A (18), and of JNK1 in complex with the KIM sequence from its scaffolding protein, JIP1 (19), have been reported. These structures identified a docking groove that binds the ΦA-X-ΦB hydrophobic motif. Interestingly, this docking groove is near to the common docking domain but is distinct from that proposed based on mutagenesis data (14). This observation raises the question of whether electrostatic interactions are important for docking or whether there exist alternative modes of binding between KIM sequences and MAP kinases.
We investigated the molecular basis of ERK2 recognition by MKP3 (13, 16, 20, 21), an ERK-specific dual-specificity phosphatase (22, 23). MKP3 preferentially recognizes and dephosphorylates ERK2 with a kcat/Km value that is near the diffusion limit (21, 24). Mutagenesis experiments suggest that the exquisite specificity of MKP3 for ERK2 is achieved by a bipartite protein–protein interaction in which the KIM sequence in MKP3 may directly bind the docking site for high-affinity association, whereas the interaction of the ERK2 substrate binding region with other elements in MKP3 may provide additional contacts crucial for MKP3 activity and/or specificity (16, 21, 24). In the current study we have solved the crystal structure of ERK2 in complex with the KIM derived from MKP3. This structure reveals the molecular mechanism underlying the docking interaction between MKP3 and ERK2. This docking site is shared by all MAP kinases and is composed of a highly acidic patch and a hydrophobic groove, which engage the basic and the ΦA-X-ΦB residues, respectively, in the KIM sequences.
Results and Discussion
Overall Structure of ERK2–KIMMKP3 Complex.
We previously showed that the KIM (residues 60–76) in MKP3 plays a major role in high-affinity ERK2 binding and that the KIM peptide from MKP3 (KIMMKP3, Ac-60GIMLRRLQKGNLPVRAL76-NH2) binds ERK2 with a Kd value of 44 μM (13). KIMMKP3 contains both basic residues (Arg-64 and Arg-65) and the ΦA-X-ΦB motif (Leu-71–Pro-72–Val-73). To uncover the structural basis of docking interactions between ERK2 and MKP3, we sought to determine the crystal structure of ERK2 in complex with KIMMKP3. We obtained crystals of the ERK2–KIMMKP3 complex under conditions similar to those described for ERK2 (25), the only difference being the identity and concentration of the salt. The crystal structure of the complex was solved by molecular replacement using the structure of phospho-ERK2 (ref. 26; Protein Data Bank ID code 2ERK) as a search model. The structure has been refined to 2.5-Å resolution with an overall Rfree value of 26.6%. Data collection and refinement statistics are given in Table 2.
Table 2.
Data collection and refinement statistics
| Space group | P21 |
|---|---|
| Unit cell dimensions | |
| a, Å | 57.4 |
| b, Å | 67.5 |
| c, Å | 86.6 |
| β, ° | 99.5 |
| Resolution, Å | 50.0–2.5 |
| No. of unit reflections | 20,430 |
| Completeness, % (last shell) | 89.8 (71.7) |
| Redundancy | 3.12 |
| Rmerge (last shell) | 8.7 (29.8) |
| Rwork/Rfree | 17.4/26.6 |
| Non-H protein atoms | 5,581 |
| Waters | 488 |
| No. of reflections in refinement (F > 1.5σ) | 19,191 |
| rms deviation in bond length, Å | 0.005 |
| rms deviation in bond angles, ° | 1.2 |
| Average B values, Å2 | 22.5 |
Rmerge = Σhkl[(Σj|Ij − 〈I〉|)/Σj|Ij|] for equivalent observations. Rwork = Σhkl‖Fo| − |Fc‖/Σhkl|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is calculated for randomly selected 2.4% of the reflections that were omitted from the refinement.
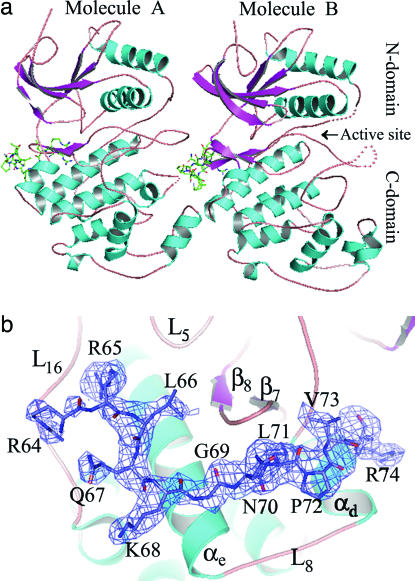
The crystallographic asymmetric unit contains two molecules (A and B) of ERK2–KIMMKP3, in which the KIM peptide is associated with the C-terminal domain of ERK2 (Fig. 1a). In both A and B, 11 of the KIM residues (R64RLQKGNLPVR74) are clearly visible in the electron-density map (Fig. 1b). The overall structures of ERK2 in molecules A and B are similar. The two structures overlap with one another with an rms deviation between all Cα atoms of 0.89 Å. The major differences between A and B lie in the phosphorylation lip (residues 172–185), the MAP kinase insert (residues 245–277), and loop195–205. For example, in molecule A the lip is completely refolded whereas in molecule B it is disordered (Fig. 1a). There are also variations in the specific interactions between the KIM peptide and ERK2 in molecules A and B as detailed below. A superposition of the ERK2 structures in the ERK2–KIMMKP3 complexes with those of the unphosphorylated (25) and phosphorylated ERK2 (26) also reveals very similar overall conformations. The rms deviations for all Cα atoms between ERK2 and A and B are 1.2 Å and 0.82 Å, respectively, whereas the rms deviations for all Cα atoms between the phospho-ERK2 and A and B are 0.59 and 0.74 Å, respectively. Again, the most notable differences between the various forms of ERK2 are in the phosphorylation lip, the MAP kinase insert, and loop195–205. Interestingly, these regions also undergo significant conformational changes when ERK2 is phosphorylated (26). It appears that these surface loops are highly flexible, although the linkage between KIM peptide binding and conformational changes in these surface loops is not apparent.
Fig. 1.
Overall structure of ERK2 bound with KIMMKP3. (a) ERK2 is shown in a ribbon model, and the disordered regions are depicted as dotted lines. The bound peptide is shown in a stick-and-ball model. (b) The Fo − Fc density map for the KIM peptide calculated by using the final model without the peptide and contoured at 2.5σ.
The KIM Docking Site in ERK2.
Complex formation between KIMMKP3 and ERK2 buries a total surface area of ≈1,250 Å2 in ERK2. Electron density for the associated KIM peptide is clear from Arg-64 to Arg-74, which include both the basic residues (Arg-64 and Arg-65) and the ΦA-X-ΦB motif (Leu-71–Pro-72–Val-73). In general, the interactions between the KIM peptide and ERK2 in molecules A and B are very similar. The KIMMKP3 peptide binds ERK2 in an extended conformation covering a contiguous surface area (the docking site) situated on the opposite of the ERK2 active site (Fig. 2a). The docking site for KIMMKP3 is nested in the C-terminal domain between L16, L5, αe, β7–β8, and αd and consists of a highly acidic patch (residues Glu-79, Tyr-126, Asp-160, Asp-316, and Asp-319) and a hydrophobic groove (residues Thr-108, Leu-110, Leu-113, Leu-119, Phe-127, and Leu-155) (Figs. 1b and 2a). Whereas docking interactions observed in previous structures of p38α–KIMMKK3b, p38α–KIMMEF2A, and JNK1–KIMJIP1 were limited to the hydrophobic groove and the ΦA-X-ΦB motif (18, 19), the structure of ERK2–KIMMKP3 reveals that KIMMKP3 interacts with the ERK2 docking site through a bipartite mechanism in which the acidic patch directly binds the basic residues and the hydrophobic groove contacts the ΦA-X-ΦB motif.
Fig. 2.
Detailed interactions between ERK2 and KIMMKP3. (a) Surface representation of ERK2 in complex with KIMMKP3, colored by electrostatic potential. KIM residues are in black, and ERK2 residues are in light blue. (b) Stereo diagram of the docking interactions between ERK2 (orange) and KIMMKP3 (green). ERK2 residues are shown in black, and those of KIMMKP3 are shown in green.
Docking Interactions Between the ΦA-X-ΦB Motif and the Hydrophobic Groove.
The ΦA-X-ΦB motif Leu-71–Pro-72–Val-73 binds to a hydrophobic groove defined by several residues in αd, αe, and the β7–β8 reverse turn (Fig. 2). The side chain of the ΦA residue Leu-71 plugs into a hydrophobic pocket surrounded by the side chains of Thr-108, Leu-113, Leu-119, Phe-127, and Leu-155. Consistent with these observations, substitution of Leu-71 in MKP3 with an Ala reduced the binding affinity of MKP3 for ERK2 by 2.6-fold (13), whereas replacement of Leu-119 in ERK2 with an Ala caused a 5.5-fold decrease in MKP3 binding affinity (16). In addition, the main-chain amide and carbonyl atoms of Leu-71 also make polar interactions with the main chain carbonyl and side chain hydroxyl of Thr-157. The X residue in the ΦA-X-ΦB motif, Pro-72, has only one H-bond through its main chain carbonyl with the side chain of Gln-117 via a water molecule, consistent with the fact that X is highly variable among proteins containing the KIM sequence. The ΦB residue Val-73 makes van der Waals contacts with the side chains of Glu-107, Thr-108, and Thr-157. Additional interactions are also observed between residues adjacent to the ΦA-X-ΦB motif and the hydrophobic groove. For example, a polar interaction exists between the side chain of Asn-70 (ΦA-1) and oxygen atom of the backbone carbonyl of Thr-157. The side chain of Arg-74 (ΦB + 1) forms both a polar contact and nonpolar contacts with the backbone carbonyl and the side chain of Lys-112, respectively. In support of these findings, elimination of the side chain at position 74 (MKP3/R74A) led to a 3.4-fold decrease in binding affinity between MKP3 and ERK2 (13).
Docking Interactions Between the Basic Residues and the Acidic Patch.
The basic residues in KIM and the common docking domain (residues 311–324) in ERK2 are known to be required for efficient interactions between the MAP kinases and their cognate binding proteins (5–10, 12, 13, 16). The structure of ERK2–KIMMKP3 provides direct evidence that Arg-64 and Arg-65 in KIMMKP3 interact with Asp-316 and Asp-319 in the common docking domain. Furthermore, the structure also reveals additional residues in ERK2 (Glu-79, Tyr-126, and Asp-160) that, together with Asp-316 and Asp-319, form a highly acidic patch to engage the basic residues as well as residues immediately C-terminal to them (Fig. 2a).
As shown in Fig. 2b, the Nφ1 atom of Arg-64 side chain mediates two hydrogen bonds, one with the carboxylate of Asp-316 and the other with the carboxylate of Asp-319. A total of four hydrogen bonds are observed between the guanidinium group Arg-65 and Asp-319 and Glu-79. In molecule A, the carboxylate of Asp-319 makes bifurcated hydrogen bonds with the Nφ1 and Nφ2 of Arg-65, whereas the Oε1 atom of the side chain of Glu-79 forms two hydrogen bonds with the Nφ1 and Nε atoms of Arg-65. In molecule B, two of the hydrogen bonds are between the side chain of Asp-319 and the Nφ1 atom of Arg-65, and the remaining two are between the Nφ1 and Nε atoms of Arg-65 and two water molecules coordinated by the side chains of Glu-79, Arg-133, and Asp-319. The observed docking interactions between the basic residues in KIMMKP3 and the acidic patch in ERK2 are in full agreement with previous mutagenesis results (13, 16). Thus, when Arg-64 or Arg-65 were replaced by an Ala, the affinity of MKP3/R64A and MKP3/R65A for ERK2 decreased 7.3- and 150-fold, respectively (13). Interestingly, the Kd value of MKP3/R64K for ERK2 was identical to that of the wild-type MKP3, indicating that a Lys can effectively replace an Arg at position 64. In contrast, MKP3/R65K exhibited a binding affinity for ERK2 that was only 3-fold better than that of MKP3/R65A, suggesting that a guanidinium side chain is required at position 65 of MKP3 for high-affinity binding with ERK2. Replacement of Asp-316 in ERK2 by an Asn resulted in a 9.6-fold decrease in MKP3 binding, whereas the affinity of ERK2/D319A for MKP3 was 106-fold lower than that of the wild-type ERK2 (16). Finally, substitution of Glu-79 or Arg-133 with an Ala reduced the affinity of ERK2/E79A and ERK2/R133A for MKP3 by 5.7- and 5.1-fold, respectively (16). These findings support the structural observations that Arg-65 in MKP3 and Asp-319 in ERK2 provide the vast majority of the side chain contacts in the ERK2–MKP3 complex.
In addition to Arg-64 and Arg-65, residues in the acidic patch also interact with Leu-66, Gln-67, and Lys-68 in KIMMKP3 (Fig. 2b). For example, the side chain of Leu-66 makes van der Waals contacts with the side chains of Thr-158 and Asp-160 in molecule A, whereas in molecule B Leu-66 makes nonpolar interactions with the side chain of Asp-160 and polar interactions with water molecules mediated by the carboxylate side chains of Asp-160 and Glu-79. Interestingly, ERK2/D160N represents a gain-of-function mutation in the Drosophila MAP kinase, termed rlSu14 (27), and the affinity of ERK2/D160N for MKP3 is 18-fold lower than that of the wild-type ERK2 (16). It is not clear from the structure how the Asp-160-to-Asn mutation affects its interactions with Leu-66 in the KIM peptide. However, because Asp-160 is part of the acidic patch, it is possible that this mutation may weaken the overall electrostatic interactions between the acidic patch and the basic residues in the KIM peptide. Finally, the main-chain carbonyls of Gln-67 and Lys-68 hydrogen-bond to two water molecules stabilized by the side chain of Asp-122 in molecule A, whereas in molecule B the carbonyl oxygen of Gln-67 hydrogen-bonds to the hydroxyl group in Tyr-126 and the carbonyl oxygen of Lys-68 interacts with the side chain of Asp-122 indirectly by means of a water molecule. Interestingly, removal of the side chain at position 126 resulted in a 20-fold reduction in MKP3 binding affinity by ERK2/Y126A (16).
Arg-65–Asp-319 Constitutes a “Hot Spot” Interaction Between MKP3 and ERK2.
Our current structural analysis and previous biochemical studies indicate that the most important residue in the ERK2 docking site for MKP3 is Asp-319, whereas the most important residue in KIMMKP3 is Arg-65. The importance of Asp-319 is underscored by the fact that it is mutated to Asn in the sevenmaker mutant of Drosophila ERK/Rolled, which displays a gain-of-function phenotype, activating several developmental pathways (28). Interestingly, the affinity of ERK2/D319N for MKP3 is 87-fold lower than that of the wild-type ERK2 (13), and ERK2/D319N is resistant to inactivation by MKPs in a number of transfected cells (29, 30). Based on the facts that removal of the side chains at Arg-65 or Asp-319 results in a similar loss in binding affinity, that Lys fails to substitute for Arg-65, and that Asn is unable to replace Asp-319, we previously proposed that the guanidinium group of Arg-65 may engage in a bidentate H-bond with the carboxylate of Asp-319, contributing to an energetic hot spot in the MKP3–ERK2 binding interface (16). This prediction has been borne out by the structure of ERK2–KIMMKP3. The structure also highlights the importance of optimal positioning and geometric complementarity between the side chains of Arg-65 and Asp-319, explaining why a large decrease in binding affinity was experienced by the conservative mutant ERK2/D319E and why swapping Arg-65 and Asp-319 simultaneously failed to restore binding affinity (16).
MAP Kinases Share a Common Docking Site.
We have uncovered the docking site for KIMMKP3, which consists of a highly acidic patch defined by several residues in αe, L16, and L5, and a hydrophobic groove between αd, αe, and the β7–β8 reverse turn. Superposition of the structure of ERK2–KIMMKP3 with those of p38α–KIMMEF2A, p38α–KIMMKK3b (18), and JNK1–KIMJIP1 (19) shows that the ΦA-X-ΦB motif in the KIM sequences bind to the same hydrophobic groove (Fig. 3). Examination of the structures suggests that a similar acidic patch also exists in all MAP kinases. This conclusion is supported by the facts that many of the residues in the acidic patch are conserved and that basic residues are present in all KIM sequences (Table 1), which are required for high-affinity binding to MKP kinases (5–13). It is likely that MAP kinases share a common docking site for KIM and that similar interactions observed between KIMMKP3 and ERK2 will be found in all MAP kinases.
Fig. 3.
Stereoview of the superposition of the structures of ERK2, p38α, and JNK1 with the bound KIM peptides highlighted (blue for KIMMKP3, green for KIMMKK3b, red for KIMMEF2A, and orange for KIMJIP1). Residues Arg-64 and Arg-65 of KIMMKP3, Arg-156 of KIMJIP1, and the ΦA and ΦB in all peptides are shown.
Potential Determinants for Docking Specificity.
If the KIMs from various MAP kinase substrates and regulators occupy the same docking site, how is specificity maintained by individual MAP kinases? To address this question, we compared the structural features of the docking sites in ERK2, p38α, and JNK1. Fig. 4depicts the acidic patch and hydrophobic groove in ERK2 superimposed on the structures of p38α and JNK1. Table 3shows a sequence alignment of the acidic patch and hydrophobic groove and highlights residues that are involved in docking interactions with the KIM sequences. As one can see, although some residues are common to all docking sites, many are unique to each MAP kinase. For example, with the exception of Leu-119 in ERK2, none of the residues lining the hydrophobic groove are identical among the MAP kinases (Fig. 4a and Table 3). Similarly, Asp-316 in ERK2 is invariant, but the remaining acidic patch residues are not strictly conserved (Fig. 4b and Table 3). These structural differences in the docking sites may impart KIM binding specificity.
Fig. 4.
Structural comparison of the ERK2, p38α, and JNK1 docking sites. (a) Structural comparison of the hydrophobic groove in ERK2 (green), p38α (orange), and JNK1 (cyan). ERK2 residues that contribute to hydrophobic interactions with the ΦA and ΦB residues are shown. (b) Structural comparison of the acidic patch in ERK2 (green), p38α (orange), and JNK1 (cyan). ERK2 residues that interact directly with the basic residues are shown.
Table 3.
Sequence alignments of the hydrophobic groove and the acidic patch in ERK2, p38α, and JNK1
| MAP kinase | Sequence |
|---|---|
| Hydrophobic groove | |
| ERK2 | 107ETDLYKLLKTQHLSNDHICYFLY129–––155LNTTCD160 |
| p38α | 110GADLNNIVKCQKLTDDHVQFLIY132–––158VNEDCE163 |
| JNK1 | 112DANLCQVIQME–LDHERMSYLLY133–––159VKSDCT164 |
| Acidic patch | |
| ERK2 | 77RHENI81––121NDHICYFLYQILR133––157TTCD160––315YDPSDE320 |
| p38α | 79KHENV83––124DDHVQFLIYQILR136––160EDCE163––312HDPDDE317 |
| JNK1 | 81NHKNI85––125HERMSYLLYQMLC137––161SDCT164––325YDPSEA330 |
Residues involved in binding the KIM peptides are shown in bold.
It appears that KIM binding specificity could be achieved by a combinatorial use of both common and unique residues in the docking site. For example, in the ERK2–KIMMKP3 structure, ΦA makes nonpolar contacts with Thr-108, Leu-113, Leu-119, Phe-127, and Leu-155. However, only three of the five corresponding residues in p38α (Ile-116, Leu-122, and Val-158) and JNK1 (Leu-123, Leu-131, and Val-159) (Table 3) are directly involved in binding ΦA of KIMMEF2A and KIMJIP1 (18, 19). Additional interactions with the ΦA residues involve Cys-162 and Cys-163, respectively, in p38α and JNK1. Although the corresponding Cys-159 is conserved in ERK2, it does not interact with KIMMKP3. Similar variations in the specific interactions are also observed between the hydrophobic grooves and the ΦB residues. Residues that are important for binding ΦB in ERK2 include Glu-107, Thr-108, Leu-113, and Thr-157, whereas only two of the corresponding residues (Ala-111 and Ile-116 in p38α and Ala-113 and Val-118 in JNK1) contact ΦB in p38α and JNK1, with additional interactions with ΦB contributed by Val-158 in p38α and Leu-115 and Met-121 in JNK1 (18, 19).
Although ERK2 and p38α share a number of residues (Glu-79, Asp-122, Tyr-128, Arg-133, Asp-316, and Asp-319) in the acidic patch, they have major differences in the sequence 157TTCD160 (160EDCE163 in p38α). Remarkably, an exchange of only two residues in this region (Thr-157 and Thr-158 in ERK2 and Glu-160 and Asp-161 in p38) converts their docking specificity for MAP kinase-activated protein kinases (14). Residues in the acidic patch of JNK1 are more diverse than those found in ERK2 and p38α (Table 3). Most notably are Glu-79 and Arg-133 in ERK2, which furnish important polar interactions with Arg-65 in KIMMKP3 (Fig. 2). The corresponding residues in JNK1 are Lys-83 and Cys-137. Obviously, the binding mode between the acidic patch in JNK1 and the basic residues in KIMJIP1 will be different from that observed in the ERK2–KIMMKP3 complex. For example, although Glu-329 of JNK1 still makes a bidentate salt bridge with Arg-156 in KIMJIP1 (ref. 19 and Fig. 3), its position is shifted ≈5 Å from that of Asp-319 in ERK2 (Fig. 4b).
Thus, although ERK2, p38α, and JNK1 employ a common docking site for KIMs, there are significant variations in the residues that are in direct contact with the peptides to ensure a high degree of binding specificity. These structural differences could be exploited to design small-molecule inhibitors that specifically target the docking site as novel reagents to modulate MAP kinase signal pathways and as starting points for potential therapeutic development (31). Finally, it is important to note that interactions between the docking site and the KIM sequence alone may be only partially responsible for recognition of effector molecules by the MAP kinases (12, 16). The role of the KIM/docking site interaction is to increase the “effective concentration” of the interacting molecules (21, 32). In addition to this “tethering” effect, additional interactions between the MAP kinases and their regulators and substrates (e.g., phosphorylation lip with MAP kinase/ERK kinases and MKPs, and substrate binding region with the phosphorylation sequence in the substrates) are required to confer the exquisite specificity in MAP kinase signaling (16).
In summary, we have solved the structure of ERK2 in complex with the KIM of MKP3. This structure reveals a KIM docking site that is comprised of a highly acidic patch and a hydrophobic groove located in a noncatalytic region opposite of the kinase active site, which mediate interactions with the basic and ΦA-X-ΦB residues, respectively, in the KIM sequences. The structure consolidates all existing biochemical data, and structural comparison indicates that this docking site is shared by all MAP kinases. Docking specificity with KIM-containing MAP kinase regulators and substrates is likely determined by both variations in the sequence and composition of the docking sites and KIMs and by the combinatorial use of both common and unique residues in the binding sites.
Materials and Methods
Peptide Synthesis, ERK2 Expression, and Purification.
The KIM peptide from MKP3 (KIMMKP3, Ac-60GIMLRRLQKGNLPVRAL76-NH2) was synthesized by using a standard protocol, purified by HPLC, and characterized by MALDI-TOF mass spectrometry by Alpha Diagnostic International. The NpT7–5 vector designed to express ERK2 with (His)6 at its N terminus was a generous gift from Elizabeth Goldsmith (University of Texas Southwestern Medical Center, Dallas) (25). The (His)6-tagged ERK2 was expressed in Escherichia coli BL21/DE3 and purified by using standard procedures of Ni2+-NTA agarose (Qiagen) affinity purification as described (13, 25).
Crystallization of the ERK2–KIMMKP3 Complex.
Initial crystallization conditions were identified by PEG/Ion screening (Hampton Research, Riverside, CA). KIMMKP3 peptide (20 μl of 20 mM stock) was added to 80 μl of 7 mg/ml ERK2 in the storage buffer (0.1 M NaCl/0.02 M Tris-HCl, pH 7.5/2 mM DTT). Crystals were obtained by hanging drop vapor diffusion by mixing 2 μl of protein solution with an equal volume of a reservoir solution containing 20% (wt/vol) PEG 3350 (Hampton Research), 0.1≈0.2 M ammonium chloride, and 100 mM 2-morpholinoethanesulfonic acid (pH 6.5). Crystals appeared in 1 week and typically reached dimensions of 0.3 mm × 0.3 mm × 0.06 mm in 2≈3 weeks. The crystals belong to space group P21 with two molecules of ERK2 and MKP3 peptide per asymmetric unit. Unit cell dimensions are as follows: a = 57.4 Å, b = 67.5 Å, c = 86.6 Å, and β = 99.6°. The solvent content of the crystal is 37%. All crystals were cryoprotected in the reservoir solution supplemented with 30% PEG 3350 and 2 mM DTT before data collection. Crystals were mounted on cryoloops (Hampton Research) and frozen in liquid nitrogen.
X-Ray Data Collection, Structure Determination, and Refinement.
X-ray data for the ERK2–KIMMKP3 crystals were collected on a Rigaku RU-300 rotating anode generator (Rigaku/Molecular Structure Corporation) equipped with focusing mirrors (Molecular Structure Corporation/Yale) and an R-AXIS IV++ image plate detector. The x-ray diffraction data extended to 2.5 Å. Images were indexed and integrated with the program denzo, and data were scaled by using scalepack (33). The data set was 89.8% complete between 50 and 2.5 Å (Rsym = 8.7%, average redundancy = 3.4, 〈I/σ〉 = 11.0). Data in the outermost shell, from 2.6 to 2.5 Å, was complete to 71.7% (Rsym = 29.8%, 2,011 unique reflections, 〈I/σ〉 = 3.2). Starting phases for ERK2–KIMMKP3 were obtained by molecular replacement by using coordinates of phosphorylated ERK2 (31) as the search model using program amore (34). The model was built into electron-density maps calculated with coefficients 2Fobs − Fcalc with the program o (35). Cycles of refinement were carried out in cns (36) by using data between 6.0 and 2.5 Å. In the final model, one asymmetric unit has two molecules of ERK2–KIMMKP3 (molecules A and B). The N-terminal Ala-(His)6 tag and the first seven residues in ERK2 are not visible in both molecules A and B. Residues 176–183 and residues 329–332 in molecule B are disordered. Side chains of residues 174, 178, and 181 have weak electron density, and no density was found around residue Lys-201 in molecule A. Eleven residues of the MKP3 KIM peptide were built manually for each molecule based on the Fobs − Fcalc map, judged by reduction in both Rwork and Rfree. The present Rwork is 17.4 (Rfree = 26.6%) for 19,191 reflections with Fobs > 1.5σ in the resolution range of 50–2.5 Å. The rms deviations from ideal bond distance and bond angle are 0.005 Å and 1.2°, respectively. The average B value for all protein atoms is 22.5 Å2. The present model includes 488 water molecules.
Acknowledgments
This work was supported by National Institutes of Health Grant CA69202 and the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations
- ERK
extracellular signal-regulated protein kinase
- KIM
kinase interaction motif
- MAP
mitogen-activated protein
- MKP
MAP kinase phosphatase
- JNK
c-Jun N-terminal protein kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The coordinates for the structure of ERK2 in complex with the KIM sequence of MKP3 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2FYS).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Widmann C., Gibson S., Jarpe M. B., Johnson G. L. Physiol. Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C., Cobb M. H. Chem. Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 3.Holland P. M., Cooper J. A. Curr. Biol. 1999;9:R329–R331. doi: 10.1016/s0960-9822(99)80205-x. [DOI] [PubMed] [Google Scholar]
- 4.Yasuda J., Whitmarsh A. J., Cavanagh J., Sharma M., Davis R. J. Mol. Cell. Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulido R., Zuniga A., Ullrich A. EMBO J. 1998;17:7337–7350. doi: 10.1093/emboj/17.24.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karim F. D., Rubin G. M. Mol. Cell. 1999;3:741–750. doi: 10.1016/s1097-2765(01)80006-x. [DOI] [PubMed] [Google Scholar]
- 7.Saxena M., Williams S., Brockdorff J., Gilman J., Mustelin T. J. Biol. Chem. 1999;274:11693–11700. doi: 10.1074/jbc.274.17.11693. [DOI] [PubMed] [Google Scholar]
- 8.Gavin A.-C., Nebreda A. R. Curr. Biol. 1999;9:281–284. doi: 10.1016/s0960-9822(99)80120-1. [DOI] [PubMed] [Google Scholar]
- 9.Smith J. A., Poteet-Smith C. E., Malarkey K., Sturgill T. W. J. Biol. Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 10.Nichols A., Camps M., Gillieron C., Chabert C., Brunet A., Wilsbacher J., Cobb M., Pouyssegur J., Shaw J. P., Arkinstall S. J. Biol. Chem. 2000;275:24613–24621. doi: 10.1074/jbc.M001515200. [DOI] [PubMed] [Google Scholar]
- 11.Sharrocks A. D., Yang S. H., Galanis A. Trends Biochem. Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 12.Tanoue T., Adachi M., Moriguchi T., Nishida E. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 13.Zhou B., Wu L., Shen K., Zhang J., Lawrence D. S., Zhang Z.-Y. J. Biol. Chem. 2001;276:6506–6515. doi: 10.1074/jbc.M009753200. [DOI] [PubMed] [Google Scholar]
- 14.Tanoue T., Maeda R., Adachi M., Nishida E. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanoue T., Yamamoto T., Nishida E. J. Biol. Chem. 2002;277:22942–22949. doi: 10.1074/jbc.M202096200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., Zhou B., Zheng C.-F., Zhang Z.-Y. J. Biol. Chem. 2003;278:29901–29912. doi: 10.1074/jbc.M303909200. [DOI] [PubMed] [Google Scholar]
- 17.Bardwell A. J., Abdollahi M., Bardwell L. Biochem. J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C.-I., Xu B.-E., Akella R., Cobb M. H., Goldsmith E. J. Mol. Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 19.Heo Y. S., Kim S. K., Seo C. I., Kim Y. K., Sung B. J., Lee H. S., Lee J. I., Park S. Y., Kim J. H., Hwang K. Y., et al. EMBO J. 2004;23:2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou B., Zhang Z.-Y. J. Biol. Chem. 1999;274:35526–35534. doi: 10.1074/jbc.274.50.35526. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y., Zhang Z.-Y. J. Biol. Chem. 2001;276:32382–32391. doi: 10.1074/jbc.M103369200. [DOI] [PubMed] [Google Scholar]
- 22.Camps M., Nichols A., Arkinstall S. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- 23.Keyse S. M. Curr. Opin. Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhou B., Wang Z.-X., Zhao Y., Brautigan D. L., Zhang Z.-Y. J. Biol. Chem. 2002;277:31818–31825. doi: 10.1074/jbc.M203969200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F., Strand A., Robbins D., Cobb M. H., Goldsmith E. J. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 26.Canagarajah B. J., Khokhlatchev A., Cobb M. H., Goldsmith E. J. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 27.Lim Y.-M., Nishizawa K., Nishi Y., Tsuda L., Inoue Y. H., Nishida Y. Genetics. 1999;153:763–771. doi: 10.1093/genetics/153.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner D., Oellers N., Szabad J., Biggs W. H., III, Zipursky S. L., Hafen E. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 29.Bott C. M., Thorneycroft S. G., Marshall C. J. FEBS Lett. 1994;352:201–205. doi: 10.1016/0014-5793(94)00958-9. [DOI] [PubMed] [Google Scholar]
- 30.Chu Y., Solski P. A., Khosravi-Far R., Der C. J., Kelly K. J. Biol. Chem. 1996;271:6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 31.Hancock C. N., Macias A., Lee E. K., Yu S. Y., Mackerell A. D., Jr., Shapiro P. J. Med. Chem. 2005;48:4586–4595. doi: 10.1021/jm0501174. [DOI] [PubMed] [Google Scholar]
- 32.Huang Z., Zhou B., Zhang Z.-Y. J. Biol. Chem. 2004;279:52150–52159. doi: 10.1074/jbc.M407820200. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z. In: Data Collection and Processing. Sawyer L., Issacs N., Bailey S. W., editors. Warrington, U.K.: Science and Engineering Council/Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 34.Navaza J. Acta Crystallogr. D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 35.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard G. J. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]