Abstract
Therapies that target estrogen signaling have made a very considerable contribution to reducing mortality from breast cancer. However, resistance to tamoxifen remains a major clinical problem. Here we have used a genome-wide functional profiling approach to identify multiple genes that confer resistance or sensitivity to tamoxifen. Combining whole-genome shRNA screening with massively parallel sequencing, we have profiled the impact of more than 56,670 RNA interference reagents targeting 16,487 genes on the cellular response to tamoxifen. This screen, along with subsequent validation experiments, identifies a compendium of genes whose silencing causes tamoxifen resistance (including BAP1, CLPP, GPRC5D, NAE1, NF1, NIPBL, NSD1, RAD21, RARG, SMC3, and UBA3) and also a set of genes whose silencing causes sensitivity to this endocrine agent (C10orf72, C15orf55/NUT, EDF1, ING5, KRAS, NOC3L, PPP1R15B, RRAS2, TMPRSS2, and TPM4). Multiple individual genes, including NF1, a regulator of RAS signaling, also correlate with clinical outcome after tamoxifen treatment.
Keywords: genetics, endocrine therapy, RNA interference screening, next generation sequencing
Approximately 70% of breast tumors express estrogen receptor α (ER) (1), which binds and mediates many of the effects of the hormone estrogen. Estrogen signaling is known to modulate several processes relevant to tumorigenesis mostly by the activity of ER as a transcription factor (2). After binding estrogen ER interacts with coactivators, resulting in the regulation of histones and gene expression (3). Of particular importance are the effects of estrogen/ER on cyclin D1 (4) and c-Myc expression (5), which are likely drivers of estrogen-stimulated cellular proliferation.
The dependence of a significant proportion of breast cancers upon estrogen signaling has been studied since the 1890s (6) and culminated in the development of pharmacological agents that inhibit ER signaling, including tamoxifen (7). Tamoxifen has gone on to become the most widely used drug in managing breast cancer. However, as with many cancer treatments, resistance to tamoxifen is a significant issue, and up to 40% of early-stage breast cancer patients who receive tamoxifen as an adjuvant therapy eventually relapse with tamoxifen-resistant disease (8).
Despite intense study, the molecular alterations that underlie endocrine therapy resistance are not fully understood, and this has limited the development of effective approaches for preventing and overcoming resistance. Nevertheless, two general mechanisms have been proposed to explain the development of resistance: (i) continued ER signaling in the presence of ER antagonists or the absence of estrogen (9), and (ii) the use of non-ER pathways that circumvent the reliance upon ER signaling (10). The activity of signal-transducing kinases has been implicated in both of these mechanisms (11, 12), and considerable effort has been made to characterize the role of individual genes in endocrine therapy resistance, with the notable findings that PAK1 and AKT activation can cause resistance to tamoxifen (13, 14). However, although candidate-based studies have been informative, a complementary approach is to interrogate the entire genome to uncover potential unique mechanisms of resistance, and high-throughput RNAi screening allows such systematic analysis to be performed (15).
Here we identified multiple genes that modulate the cellular response to tamoxifen by carrying out an unbiased genome-wide functional screen, coupling an shRNA interference library to pool deconvolution by massively parallel sequencing.
Results
Tamoxifen Genome-Wide RNAi Screen.
To identify determinants of tamoxifen response, we designed a high-throughput RNAi screen (SI Appendix, Fig. S1A). In brief, the screening procedure involved (i) infecting a 4-hydroxytamoxifen (4OHT, an active metabolite of tamoxifen) sensitive, estrogen receptor positive (ER+), breast tumor cell line (MCF7) with a library encompassing 56,670 shRNA-coding lentiviral constructs designed to target 16,487 human protein-coding genes (OpenBiosystems GIPZ shRNAmir human genome-wide library), (ii) after allowing the viral constructs to integrate into the tumor cell genome, (iii) exposing cells to 4OHT for 21 d, (iv) recovering shRNA target sequences from the genomic DNA of the surviving cell fraction by PCR amplification, and then (v) estimating the frequency of each shRNA sequence in surviving cells (and thus the effect of each shRNA upon 4OHT response) by sequencing PCR amplification products by massively parallel sequencing.
Before MCF7 cell infection, we divided the genome-wide shRNA library into six pools (each pool comprising 9,600 shRNAs) and infected cells with each pool separately. Cells were infected at multiplicity of infection of 0.7, and on average 2,000 cells per shRNA construct were infected (SI Appendix, Fig. S2). Seventy-two hours after infection, cells were selected in puromycin for 2 d to remove the nontransduced fraction and then divided into two samples: one subsequently exposed to 4OHT solubilized in ethanol, the other exposed to ethanol alone. Five days after initial infection, drug exposure was initiated and sustained for 21 d to model chronic tamoxifen exposure used clinically. A final 4OHT concentration of 500 nM was used, which caused 40% inhibition of cell survival over the time course of the screen (surviving fraction 60%, SF60). To identify shRNA constructs that modulated the response to 4OHT, we estimated shRNA frequency in 4OHT and vehicle-treated cultures at the end of this 21-d period using massively parallel sequencing.
After PCR products were sequenced (SI Appendix, Table S1), each short read was matched to the corresponding RNAi target in the reference library. We then used the frequency of each individual short-read sequence to estimate the frequency of shRNAs in each surviving population. To identify shRNAs that conferred either 4OHT resistance or sensitivity effects, we compared the representation of shRNAs in the 4OHT-treated samples with those in vehicle-treated samples. First, data from each of three biological replicate screens were normalized to account for experimental intrascreen variation (variation between different viral pools). In doing this, we also corrected biases associated with differential starting representations of shRNAs. Second, we applied an interreplica screen rank normalization and used these values to ascribe each shRNA a drug effect (DE) score that represented the magnitude of 4OHT resistance (positive DE) or sensitivity (negative DE) (SI Appendix, Fig. S1B). Here overrepresentation of an shRNA in a 4OHT-treated sample indicated a resistance-causing effect, whereas underrepresentation indicated a sensitization effect.
Identification of Screen Hits.
To define 4OHT-modulating effects from the screen data, we used three parallel strategies previously used in RNA interference screens. In the first instance we selected significant effects according to the variance of the entire dataset. We calculated the median absolute deviation (MAD) to estimate the variance of the normalized data (16) and defined resistance-causing hits as those shRNAs that gave DE scores >2 × MAD (Z score >2), a threshold approximately equal to 2 SDs from the median. Sensitization effects were defined as shRNAs that returned DE scores of Z < −2. In addition to this approach we also used RNAi Gene Enrichment Ranking (RIGER) (17), as implemented in the Broad Institute's GENE-E software package. In brief, RIGER calculated the weighted sum of the two top-ranked shRNAs for each gene on the basis of the log fold change between three biological replicates for each condition, and provided a normalized enrichment score per gene. Finally, we also used RNAi Set Analysis (RSA), a modification of Gene Set Analysis (http://www-stat.stanford.edu/∼tibs/GSA/) that uses maximum–mean statistics to identify significantly enriched or depleted shRNA sets.
In total, Z score threshold identified 1,049 resistance-causing genes and 1,126 sensitization genes, RIGER generated a list of 504 resistance-causing and 498 sensitization genes with a P value of <0.05, and RSA generated a list of 443 resistance-causing genes and 477 sensitization genes with a false discovery rate approaching zero. Given the limitations of each method, we took a pragmatic approach and considered a subset of the genes identified by all three methods for further examination. This intersection approach identified 121 candidate genes mediating sensitization and 131 candidate genes mediating resistance to tamoxifen (Fig. 1A and SI Appendix, Table S2).
Fig. 1.
Detection of tamoxifen sensitization and resistance-causing effects. (A) Venn diagrams indicating the number of candidate hits defined by three parallel analysis methods. (B) Plot of shRNA DE Z scores ranked by size of effect. Data from nonsilencing shRNAs is highlighted, as are data from shRNA targeting PTEN or ESR1, the gene encoding ERα.
We examined screen performance using positive and negative controls. As expected, nonsilencing control shRNAs did not modulate the cellular response to 4OHT, whereas four shRNAs targeting ESR1, the gene encoding ERα, caused sensitivity to 4OHT (Fig. 1B). In vitro functional studies as well as clinical data suggest that reduced PTEN activity causes resistance to tamoxifen (17). Five shRNAs targeting PTEN caused 4OHT resistance in the screen (Fig. 1B), and three PTEN shRNAs chosen for further investigation caused 4OHT resistance as assessed using a GFP competition assay (SI Appendix, Fig. S3 A and C). Similarly, an siRNA pool targeting PTEN was able to recapitulate the resistance phenotype in MCF7 cells treated with either 4OHT or a pure antiestrogen, ICI182,780, using a previously validated 96-well arrayed method (15) (SI Appendix, Fig. S3 B and C). As the PTEN effect identified in the genome-wide screen was confirmed using two orthogonal assays, we reasoned that the screen itself and the analysis methods used were able to identify true endocrine therapy-modulating effects.
Validation of Screen Effects.
Despite their utility, RNAi screens are prone to artifacts such as off-target and screen format-specific effects (18). To overcome these issues, we selected genes identified in the genome-wide shRNA screen and examined them using an independent mode of RNA interference (siRNA) and a 96-well arrayed method. The target sequences for siRNAs used in this validation step did not overlap with the target sequences of shRNAs present in the initial screen (SI Appendix, Table S3), and this allowed us to minimize the impact of off-target effects.
Using this assay system and a single concentration of 4OHT, siRNA duplexes recapitulated 23 tamoxifen-modulating effects (Fig. 2A). Focusing on this 23-gene set, we established 4OHT dose–response relationships for each gene (SI Appendix, Table S4). In addition to PTEN, we validated 11 genes whose RNAi targeting caused 4OHT resistance (BAP1, CLPP, GPRC5D, NAE1, NF1, NIPBL, NSD1, RAD21, RARG, SMC3, and UBA3) (Fig. 2B) and 11 genes that when targeted caused sensitivity (ESR1, C10orf72, C15orf55/NUT, EDF1, ING5, KRAS, NOC3L, PPP1R15B, RRAS2, TMPRSS2, and TPM4) (Fig. 2C and SI Appendix, Table S5).
Fig. 2.
Validation of individual tamoxifen resistance- or sensitivity-causing effects. (A) Effect of siRNA on survival in 4OHT. MCF7s were transfected with siRNA duplexes in replica plates and 48 h later exposed to 1 μM 4OHT or drug vehicle. Seven days later cell viability was determined using Cell Titer Glo. Surviving fraction for each siRNA was calculated using the calculation SFgene x = luminescence in 4OHT-treated wells/luminescence in similarly transfected vehicle-treated wells. (B) Individual dose–response curves for 11 resistance-causing genes. Experimental setup was as in A but using a 4OHT titration. (C) Individual dose–response curves for 11 sensitivity-causing genes.
Network analysis using STRING (19) identified a number of well-established molecular associations among these validated genes (Fig. 3A). These included the identification of both components of the UBA3/NAE1 heterodimer, which mediates the conjugation of NEDD8, an ubiquitin-like protein, to substrates (20). siRNAs targeting either UBA3 or NAE1 (SI Appendix, Fig. S4), in addition to causing resistance to 4OHT, also caused resistance to ICI182,780 (21). The resistant phenotype was also reproduced by siRNA silencing of NEDD8 itself (Fig. 3B), thus validating our screen observations. Interestingly, members of this neddylation pathway, and the UBA3/NAE1 complex in particular, have previously been reported to mediate the degradation of ER (22).
Fig. 3.
Network-associated effects. (A) Interaction diagrams for selected validated hits. In each case, the type interaction is shown. (B) ICI182,780 and 4OHT dose–response curves for selected genes.
We also demonstrated that a number of cohesion-associated/chromatin remodeling proteins modulated response to both 4OHT and ICI182,780. The cohesin protein complex is involved in maintaining sister chromatid proximity in dividing cells and has recently been implicated in mediating transcriptional insulation and control via interactions with CTCF (23). siRNA targeting RAD21 (24), SMC3, and NIPBL (25), caused 4OHT and ICI182,780 resistance (Figs. 2B and 3B). The precise mechanism whereby cohesin components affect tamoxifen sensitivity is not yet clear, but it seems possible that imbalance of these proteins alters the transcriptional profile of ER+ tumor cells, enabling continued growth in the face of endocrine therapy (23).
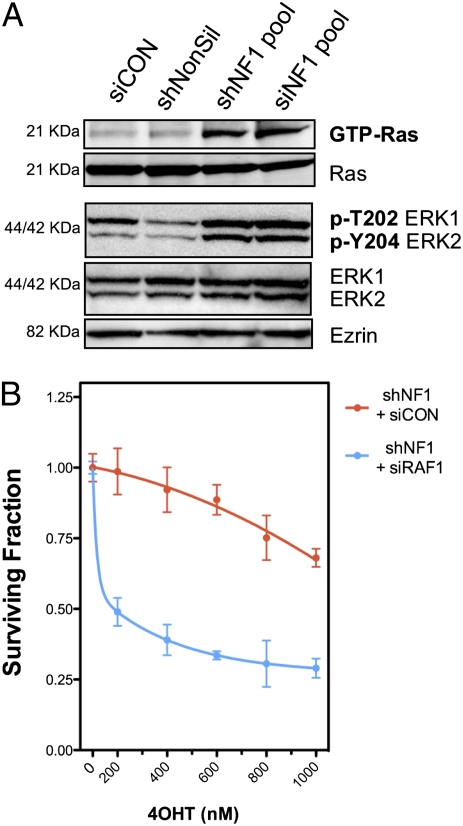
Finally, a number of proteins involved in RAS signaling were also found to modulate the response to 4OHT. Both shRNA and siRNAs targeting NF1 (neurofibromin), a GTPase tumor suppressor protein, caused 4OHT resistance (Figs. 2B and 3B). NF1 mediates the inactivation of classic RAS proteins (e.g., KRAS and HRAS) but also nonclassic RAS proteins, including RRAS2 (26). siRNAs targeting KRAS and RRAS2 both caused 4OHT sensitivity, and KRAS silencing also caused moderate ICI182,780 sensitivity (Figs. 2C and 3B), as did the downstream mediator of RAS signaling, c-RAF (RAF1) (Fig. 3B). To verify that silencing of NF1 affected RAS signaling in the face of 4OHT treatment, we measured levels of active RAS (GTP-RAS) and phosphorylated ERK in MCF7 cells treated with 4OHT, and these were indeed elevated with NF1 silencing (Fig. 4A). Furthermore, silencing of c-Raf in MCF7 cells with stable NF1 silencing alleviated tamoxifen resistance (Fig. 4B), supporting the hypothesis that these effects were, in part at least, via RAS/RAF signaling.
Fig. 4.
NF1 and tamoxifen response. (A) NF1 RNAi reagents that cause 4OHT resistance activate RAS and ERK in MCF7 cells treated with 4OHT. MCF7 cells were transfected with the indicated RNAi reagents and treated with 4OHT (500 nM for 24 h), after which levels of GTP-Ras and phosphorylated ERK were detected by immunoblotting. (B) Suppression of downstream RAS effector, c-RAF (RAF1), restores sensitivity to tamoxifen in MCF7s with stable NF1 knockdown. MCF7 cells were infected with NF1 shRNA and, after puromycin selection, subsequently transfected with nontargeting or RAF1 siRNAs. The response to 4OHT was then monitored using a 96-well format assay (15).
Clinical Significance.
Having identified a compendium of genes that modulated the response to endocrine therapy, we assessed whether they were associated with clinical response to endocrine therapy. To do this we interrogated publicly available microarray gene expression profiles from tumors of patients subsequently treated with adjuvant tamoxifen. Specifically, we used five patient datasets for this analysis: Stockholm GSE1456, n = 87 (STOCK in Fig. 5) (27); Oxford GSE6532, n = 109 (OXFT) (28); Karolinska Institute GSE3494, n = 72 (KIT) (29); Guy's Hospital GSE9195, n = 77 (GUYS77) (30); and Guy's Hospital GSE6532, n = 87 (GUYS87) (28). In each of these datasets, biopsies were taken from ER+ breast tumors before tamoxifen treatment. We obtained the normalized gene expression levels for each of the 23 functionally validated genes and compared these with a surrogate marker of tamoxifen response, the time to distant relapse. To quantify associations between gene expression and the likelihood of relapse/tamoxifen resistance, we calculated Cox proportional log hazard ratios (HRs) (31) for each gene as the relative risk of distant relapse in terms of gene expression. This analysis suggested that reduced expression of NF1, a gene whose silencing caused tamoxifen resistance in the functional screen, was associated with a statistically significant higher risk to distant relapse when considering all datasets (P < 0.05; Fig. 5A). Reduced NF1 expression did not, however, correlate with well-established prognostic factors (SI Appendix, Table S6), suggesting that it was a relatively independent marker of response to tamoxifen. A similar analysis of the validated sensitivity-causing effects indicated that reduced expression of EDF1 was associated with a statistically significant lower risk to distant relapse in a combined series of independent cohorts (P < 0.05; SI Appendix, Fig. S5).
Fig. 5.
Clinical significance of genes identified in the functional screen. (A) Low NF1 expression correlates with poor outcome in tamoxifen-treated patients. Forest plot indicates log HRs for increase in NF1 expression as estimated by Cox regression. Dots represent the weighted median effect and lines the 95% confidence interval (CI) for each of the five studies considered. Log HRs refer here to the relative risk of distant relapse in relation to gene expression, whereby positive and negative values represent decreasing or increasing risk, respectively, as gene expression decreases (zero suggests no correlation between gene expression and relative risk). A global negative log HR indicates that reduced expression of NF1 correlates with a poor outcome (high risk) in tamoxifen-treated breast cancer patients. As diamond limits define CI for the overall effect, the combined effect for NF1 is significant at P < 0.05. The most significant individual study is presented as a Kaplan-Meier survival curve in SI Appendix, Fig. S6. (B) Low expression of eight resistance-causing genes identified in the functional screen correlates with poorer outcome in tamoxifen-treated patients. Genes included in this analysis were BAP1, CDK10, NF1, NIPBL, PTEN, RARG, SMC3, and UBA3. (C) Low expression of six sensitivity-causing genes identified in the functional screen correlates with a favorable outcome in tamoxifen-treated breast cancer patients. Genes included in this analysis were EDF1, KRAS, PDPK1, RAF1, TMPRSS2, and TPM4.
Although interrogating the relationship between individual gene expression and clinical outcome parameters in tamoxifen-treated patients can provide some insight into the clinical relevance of the genetics of tamoxifen resistance, it is also possible that a constellation of individually modest gene effects combine to generate the final resistant phenotype. Given this, we tested whether aggregate measures of expression from groups or modules of genes (metagenes) also correlated with time to distant relapse in patients treated with tamoxifen. Using the same patient datasets and established methodology (32), we defined two functional metagenes for this analysis: one populated by genes shown in this or our previous study (15, 33) to cause resistance to tamoxifen, and a second similarly curated gene set defined by sensitivity-causing effects that also correlated with outcome data. In total, these modules comprised eight genes functionally predicting resistance [BAP1, RARG, PTEN, SMC3, NF1, NIPBL, UBA3, and CDK10 (15)] (Fig. 5B) and six genes functionally predicting sensitivity [EDF1, KRAS, RAF1, TMPRSS2, TPM4, and PDPK1 (33)] (Fig. 5C). This analysis suggested that both “resistance” and “sensitivity” metagenes correlated with clinical outcome (P < 0.05) and gave moderately greater predictions of outcome than the use of individual genes such as NF1.
Discussion
Here, we report a genome-wide functional screen to identify determinants of tamoxifen sensitivity. This screen and the subsequent validation experiments define a constellation of genes that modulate the cellular response to this widely used drug. Notable among the set of genes identified are multiple components of a neddylation complex, a series of genes that modulate cohesion-associated/chromatin remodeling and also multiple members of the RAS signaling cascade. Of these, the mRNA expression levels of genes such as NF1 appear to correlate with clinical outcome, as do two metagenes that encompass genes functionally shown to modulate the response to tamoxifen.
Our functional screen provides additional evidence for a key role of RAS/RAF/MEK/ERK signaling in determining the response to endocrine agents. Previously, we demonstrated that silencing of the kinase CDK10 was sufficient to cause 4OHT and ICI182,780 resistance, an effect likely mediated by the loss of CDK10 suppression on RAF1 expression and subsequent up-regulation of RAS/RAF/MEK/ERK signaling (15). Furthermore, other studies have demonstrated that transfection of constitutively active MEK1 or RAF1 cDNA expression constructs into MCF7 cells results in hyperactivation of p42/p44 MAPK and acquisition of antiestrogen resistance (34). In the genome-wide screen reported here shRNA targeting NF1 (neurofibromin), a GTPase that suppresses tumor formation by inhibiting RAS activation, caused 4OHT resistance, and RNAi targeting of KRAS caused 4OHT sensitivity, thus adding to the known nodal points within this signaling cascade that can modulate drug response. In addition, RRAS2 (TC21) silencing also resulted in tamoxifen sensitivity. RRAS2 is a RAS family member that shares more than 50% amino acid sequence identity with classic RAS proteins (HRAS, NRAS, and KRAS). RRAS2 encompasses almost identical functional domains to these latter proteins and accordingly shares a number of effectors, including RAF1 (35). A link between RRAS2 and tamoxifen response has already been suggested. Specifically, high RRAS2 protein expression in breast tumor biopsies has been shown to correlate with a shorter time to relapse in tamoxifen-treated patients, as does possession of a RRAS2 allele carrying the −582C→T RRAS2 promoter polymorphism present in ≈34% of European Caucasians (36). Our functional data support the case for RRAS2 playing a critical role in determining the response to tamoxifen and strengthens the argument for examining the functional effects of RRAS2 polymorphisms and how these relate to clinical outcome.
Genome-wide functional screens, such as that described here, should provide annotated gene lists that may act as a starting point for the in-depth dissection of clinically relevant phenotypes, in much the same way as microarray profiling has allowed an understanding of gene expression alterations associated with disease. As RNAi screening technology and reagents mature, the functional screening of selected gene subsets is now within the means of most laboratories. Furthermore, the experimental setup applied here, with the deconvolution of pooled shRNAs by massively parallel sequencing, allows genome-wide screens to be performed at a fraction of the cost and time compared with genome-wide plate arrayed screens and will likely enable the dissection of additional disease-based phenotypes.
Materials and Methods
Pooled shRNA Screen Method.
As a screening library, we used the OpenBiosystems GIPZ human genome-wide shRNA library. Lentiviral DNA was generated according to the manufacturer's instructions and then pooled to generate six pools, each containing 9,600 different shRNA constructs. Virus was packaged in 293T cells and MCF7 cells transduced with a final representation of ≈1,000 cells per shRNA (20,000,000 per shRNA pool). Transduction efficiency was estimated by GFP detection using FACS 72 h after infection. Cells were then cultured in 1 mg/L puromycin for 2 d to enrich for transduced cells and then continuously cultured in media containing 500 nM 4OHT or 0.5% (vol/vol) ethanol for 21 d. Media with drug was replenished every 2 to 3 d. MCF7 cells were cultured in phenol red-free RPMI-1640 media supplemented with 10% (vol/vol) charcoal-dextran stripped FBS. Massively parallel sequencing for shRNA retrieval and screen data analysis are described in SI Appendix, SI Methods and Materials.
Clinical Analysis.
This was performed using tumor samples from five independent datasets of ER+ patients subject to tamoxifen adjuvant therapy. Raw Affymetrix U133A microarray expression files were downloaded from the Gene Expression Omnibus repository (accession codes per study indicated in main text) and processed using the RMA function from the Bioconductor Affy package for R (37). Log–rank analyses and Kaplan-Meier plots were produced using the “survdiff” and “plot.survfit” S-plus (TIBCO Software) functions, respectively. Normalized gene expression values were partitioned into quartiles. Cox proportional hazards regression was carried out using the “coxph” S-plus function. Here we used gene expression values as the sole continuous predictor variable. Random effects metaanalysis was carried out using the β values estimated by this regression.
Supplementary Material
Acknowledgments
This work was funded by Breakthrough Breast Cancer, The Breast Cancer Research Foundation, and American Association for Cancer Research/Stand Up To Cancer. We acknowledge National Health Service funding to the Royal Marsden Hospital National Institute for Health Research Biomedical Research Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018872108/-/DCSupplemental.
References
- 1.EBCTCG Tamoxifen for early breast cancer: An overview of the randomised trials. Early Breast Cancer Trialists' Collaborative Group. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 2.Frasor J, et al. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: Insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: Involvement of a cAMP response-like element. Proc Natl Acad Sci USA. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubik D, Shiu RP. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J Biol Chem. 1988;263:12705–12708. [PubMed] [Google Scholar]
- 6.Beatson G. On the treatment of inoperable cases of carcinoma of the mammary. Suggestions for a new method of treatment with illustrative cases. Lancet. 1896;2:104–107. [PMC free article] [PubMed] [Google Scholar]
- 7.Cole MP, Jones CT, Todd ID. A new anti-oestrogenic agent in late breast cancer. An early clinical appraisal of ICI46474. Br J Cancer. 1971;25:270–275. doi: 10.1038/bjc.1971.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- 9.Shou J, et al. Mechanisms of tamoxifen resistance: Increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96:926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 10.Oh AS, et al. Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol. 2001;15:1344–1359. doi: 10.1210/mend.15.8.0678. [DOI] [PubMed] [Google Scholar]
- 11.Gee JM, Robertson JF, Ellis IO, Nicholson RI. Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. Int J Cancer. 2001;95:247–254. doi: 10.1002/1097-0215(20010720)95:4<247::aid-ijc1042>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.Pérez-Tenorio G, Stål O. Southeast Sweden Breast Cancer Group Activation of AKT/PKB in breast cancer predicts a worse outcome among endocrine treated patients. Br J Cancer. 2002;86:540–545. doi: 10.1038/sj.bjc.6600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm C, et al. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–680. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 14.Campbell RA, et al. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: A new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 15.Iorns E, et al. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell. 2008;13:91–104. doi: 10.1016/j.ccr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 17.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 19.Snel B, Lehmann G, Bork P, Huynen MA. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 21.Jones SE. A new estrogen receptor antagonist—an overview of available data. Breast Cancer Res Treat. 2002;75(Suppl 1):S19–S21. doi: 10.1023/a:1020357631871. discussion S33–S35. [DOI] [PubMed] [Google Scholar]
- 22.Fan M, Bigsby RM, Nephew KP. The NEDD8 pathway is required for proteasome-mediated degradation of human estrogen receptor (ER)-alpha and essential for the antiproliferative activity of ICI 182,780 in ERalpha-positive breast cancer cells. Mol Endocrinol. 2003;17:356–365. doi: 10.1210/me.2002-0323. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt D, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakimi MA, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, et al. Role of TC21/R-Ras2 in enhanced migration of neurofibromin-deficient Schwann cells. Oncogene. 2004;23:368–378. doi: 10.1038/sj.onc.1207075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 29.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loi S, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox DR. Regression models and life tables. J R Stat Soc, B. 1972;34:187–220. [Google Scholar]
- 32.Juul N, et al. Assessment of an RNA interference screen-derived mitotic and ceramide pathway metagene as a predictor of response to neoadjuvant paclitaxel for primary triple-negative breast cancer: A retrospective analysis of five clinical trials. Lancet Oncol. 2010;11:358–365. doi: 10.1016/S1470-2045(10)70018-8. [DOI] [PubMed] [Google Scholar]
- 33.Iorns E, Lord CJ, Ashworth A. Parallel RNAi and compound screens identify the PDK1 pathway as a target for tamoxifen sensitization. Biochem J. 2009;417:361–370. doi: 10.1042/BJ20081682. [DOI] [PubMed] [Google Scholar]
- 34.El-Ashry D, Miller DL, Kharbanda S, Lippman ME, Kern FG. Constitutive Raf-1 kinase activity in breast cancer cells induces both estrogen-independent growth and apoptosis. Oncogene. 1997;15:423–435. doi: 10.1038/sj.onc.1201198. [DOI] [PubMed] [Google Scholar]
- 35.Calvo F, Crespo P. Structural and spatial determinants regulating TC21 activation by RasGRF family nucleotide exchange factors. Mol Biol Cell. 2009;20:4289–4302. doi: 10.1091/mbc.E09-03-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rokavec M, et al. A polymorphism in the TC21 promoter associates with an unfavorable tamoxifen treatment outcome in breast cancer. Cancer Res. 2008;68:9799–9808. doi: 10.1158/0008-5472.CAN-08-0247. [DOI] [PubMed] [Google Scholar]
- 37.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.