Abstract
The clinical efficacy of EGFR kinase inhibitors is limited by the development of drug resistance. The irreversible EGFR kinase inhibitor WZ4002 is effective against the most common mechanism of drug resistance mediated by the EGFR T790M mutation. Here we show, in multiple complementary models, that resistance to WZ4002 develops through aberrant activation of ERK signaling caused by either an amplification of MAPK1 or by downregulation of negative regulators of ERK signaling. Inhibition of MEK or ERK restores sensitivity to WZ4002 and prevents the emergence of drug resistance. We further identify MAPK1 amplification in an erlotinib resistant EGFR mutant NSCLC patient. In addition, the WZ4002 resistant MAPK1 amplified cells also demonstrate an increase both in EGFR internalization and a decrease in sensitivity to cytotoxic chemotherapy. Our findings provide insights into mechanisms of drug resistance to EGFR kinase inhibitors and highlight rationale combination therapies that should be evaluated in clinical trials.
Keywords: Drug resistance, EGFR mutation, gene amplification
Introduction
Epidermal growth factor receptor (EGFR) kinase inhibitors gefitinib and erlotinib are effective clinical therapies for non-small cell lung cancer patients harboring EGFR mutant cancers. Several phase III clinical trials have demonstrated improved clinical efficacy compared to systemic chemotherapy (1–3). However, despite these benefits, all patients ultimately develop acquired resistance to gefitinib and erlotinib (4). The most common mechanism, detected in 50–60% of patients, of acquired resistance is mediated by the secondary EGFR T790M mutation, and results in an increase in ATP affinity (5–8). In preclinical models, irreversible quinazoline based EGFR inhibitors, including afatinib (BIBW2992) and dacomitinib (PF299804), effectively inhibit the growth of EGFR T790M containing cell line models in vitro (9, 10). The covalent binding allows these inhibitors to achieve greater occupancy of the ATP-site relative to the gefitinib or erlotinib, thus providing the ability to inhibit EGFR T790M (8). However, in clinical studies, afatinib did not prolong survival compared to placebo in NSCLC patients that had developed acquired resistance to gefitinib or erlotinib (11). Furthermore, in preclinical studies, resistance of EGFR T790M tumor cells to dacomitinib develops rapidly and is caused by amplification of the T790M containing allele (12).
In an effort to overcome the therapeutic limitations of irreversible quinazoline EGFR inhibitors, we previously identified a novel class of irreversible pyrimidine-based EGFR kinase inhibitors (13). These agents, including WZ4002, are more potent than irreversible quinazoline EGFR inhibitors in EGFR T790M bearing models, but are less potent inhibitors of wild type (WT) EGFR (13). Coupled with the increased potency, the mutant selective property of this class of agents may provide the ability to achieve clinical concentrations sufficient to inhibit EGFR T790M.
In the current study we modeled acquired resistance to WZ4002 in EGFR T790M containing models in vitro and in vivo. We undertook these studies in an effort to identify potential strategies that can be used to enhance the efficacy of this class of EGFR inhibitors. Our studies identify a novel mechanism of resistance to EGFR inhibitors and inform the development of a novel combination therapeutic strategy that can be used to effectively treat EGFR T790M containing cancers.
Results
WZ4002 resistant cells contain an amplification in MAPK1
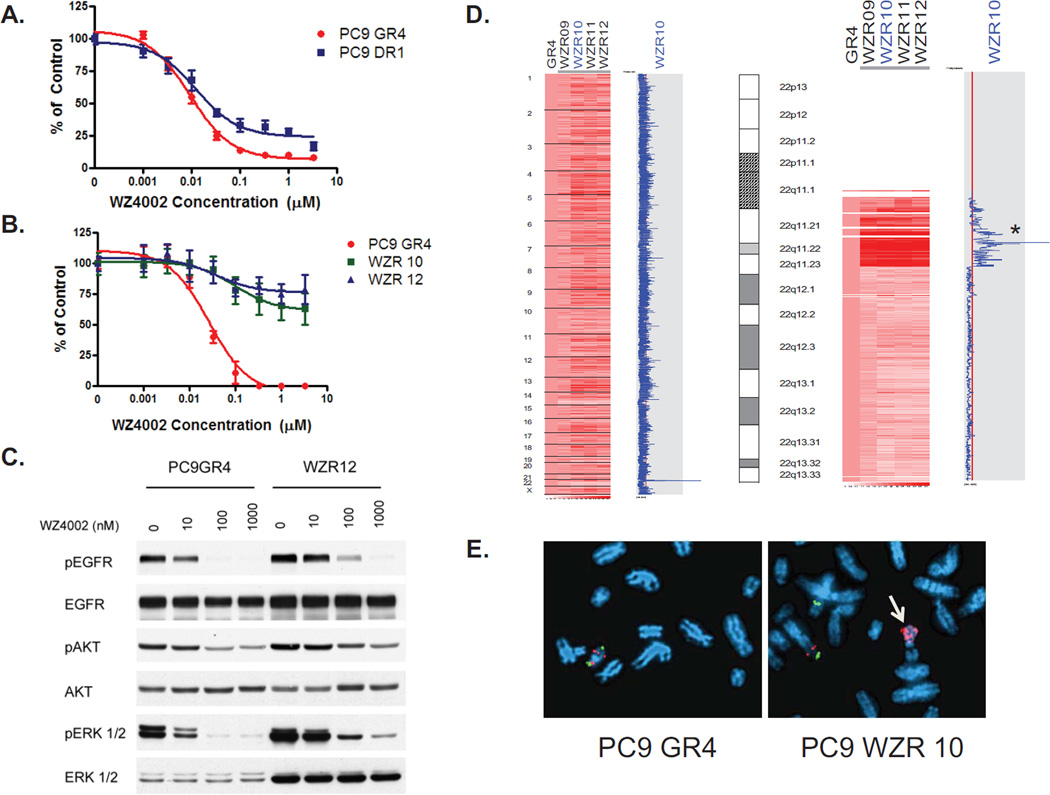
In our prior studies we generated gefitinib resistant (GR) version of the EGFR mutant PC9 (Del E746_A750) cell line (13). These cells contain the EGFR T790M resistance mutation and are sensitive to WZ4002 (13). When we exposed the PC9 GR cells to dacomitinib (PF299804), a clinical irreversible quinazoline EGFR inhibitor and generated resistant cells, they contained a focal amplification in EGFR preferentially involving the T790M allele (12). These PC9 DR (dacomitinib resistant) cells are as sensitive to WZ4002 as the parental PC9 GR cells (Fig. 1A). In order to determine how cancers that harbor an EGFR T790M develop resistance to WZ4002, we generated WZ4002 resistant (WZR) versions of the PC9 GR4 cells using previously established methods (12, 14). Several individual resistant clones were identified and confirmed to be drug resistant (Fig 1B). The resistant cells still harbored the EGFR DelE746_A750/T790M double mutation but contained no additional EGFR mutations (data not shown) and were also cross resistant to dacomitnib and afatinib (data not shown). WZ4002 still inhibited EGFR phosphorylation in the resistant cells, although slightly less potently in the GR4 cells, but more noticeably, this inhibition was decoupled from inhibition of downstream signaling most notably ERK2 phosphorylation (Fig. 1C). The WZR12 cells contain higher levels of both total and phosphorylated ERK2 than the PC9 GR cells (Fig 1C). In order to determine whether there was a genomic basis for the increase in ERK2 protein, we performed a genome wide copy number analysis of the WZ resistant cells and compared them to the parental PC9 GR4 cells (Fig. 1D). The WZR cells contain an amplification in chromosome 22 which is not present in the parental drug sensitive cell line. This region contains the gene, MAPK1, which encodes ERK2 (Fig 1D). We confirmed the MAPK1 amplification using both fluorescence in situ hybridization (FISH) (Fig 1E.) and quantitative PCR (Fig. S1). The amplification also led to increased MAPK1 gene expression (Fig S2A).
Figure 1.
WZ4002 resistant EGFR mutant Del E746_A750/T790M cells contain an amplification in MAPK1. A. PC9 GR4 (Del E746_A750/T790M) and DR1 (Del E746_A750/T790M amplification) cells were treated with WZ4002 at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. B. WZR10 and WZR12 cells are resistant to WZ4002. Cells were treated with WZ4002 as in A. C. PC9GR4 and WZR12 cells were treated with WZ4002 at indicated concentrations for 6 hours. Cell extracts were immunoblotted to detect the indicated proteins. D. WZR cells contain an amplification in MAPK1. The PC9 WZR clones (right) were compared with the PC9 GR4 cells (first column). The blue curve on the right indicates degree of amplification of each SNP from 0 (left) to 8 (right). Left, genome wide view; Right, detailed view of chromosome 22. The genomic location of MAPK1 is indicated by an asterix. E. Metaphase FISH of PC9 GR4 and WZR10 cells using MAPK1 (red) and reference probe (green; RP11-768L22). Amplification of MAPK1 is observed in WZR10 cells (arrow).
Inhibition of MAPK signaling restores sensitivity to WZ4002
We next evaluated whether inhibition of MAPK signaling would restore sensitivity to WZ4002 in the PC9 WZR cells. We first determined the concentration of the MEK inhibitor CI-1040 necessary to downregulate ERK 1/2 phosphorylation to similar levels as in the parental cell lines (Fig. 2A). When used in combination with WZ4002, the MEK inhibitors CI-1040 (Fig. 2B) or GSK1120212 (Fig. S3) completely restored the sensitivity to WZ4002 in the WZR cells similar to that of the parental PC9 GR4 cells. Similarly, the combination led to complete inhibition of ERK 1/2 phosphorylation and restored WZ4002 mediated apoptosis analogous to that observed in the parental PC9GR cells (Figs. 2C and S4). WZ4002 sensitivity was also restored following downregulation of MAPK1 using a MAPK1 specific short hairpin (sh)-RNA (Figs. S5 and 2D), or when WZ4002 was combined with an ERK 1/2 kinase inhibitor (compound 11e) (Fig 2D) (15). Inhibition of ERK 1/2 using compound 11e also restored WZ4002 mediated apoptosis in the PC9 WZR cells (Figs S4). As a pharmacodynamic measure of compound 11e, we evaluated p90RSK phosphorylation, a known ERK substrate (16). In the PC9 GR cells, but not in the WZR cells, WZ4002 treatment inhibited p90RSK phosphorylation (Fig. 2E). However, in both GR4 and WZR10 cells, compound 11e was able to inhibit p90RSK phosphorylation (Fig 2F). The addition of a dual PI3K and MTOR inhibitor, PI103, or the AKT inhibitor MK-2206, did not restore sensitivity to WZ4002 nor resulted in WZ4002 mediated apoptosis (Figs. S6A–C) (17, 18).
Figure 2.
Inhibition of ERK 1/2 signaling restores sensitivity to WZ4002 in PC9 WZR cells. A. PC9 GR4 or WZR10 cells were treated with the indicated concentrations of CI-104 for 6 hrs. Cell extracts were immunoblotted to detect the indicated proteins. B. PC9GR4 or WZR10 cells were treated with WZ4002 alone at the indicated concentrations or in combination with CI-1040 (3 µM). Viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. C. PC9GR4 or WZR10 cells were treated with WZ4002 alone at indicated concentrations or with CI-1040 (3 µM) for 6 hours or 48 hours (PARP lane only). Cell extracts were immunoblotted to detect the indicated proteins. D. Cells were treated with WZ4002 at the indicated concentrations or in combination with Cmp 11E (WZR10 only). Viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. E. WZ4002 (6 hours) inhibits p90RSK phosphorylation in PC9 GR4 but not WZR10 cells. F. Cmp 11E (6 hours) inhibits p90RSK phosphorylation in both PC9 GR4 and WZR10 cells. G. WZ4002 treatment (6 hours) leads to BIM upregulation in PC9 GR4 but not WZR10 cells.
To further demonstrate the role of activated MAPK signaling in mediating WZ4002 resistance, we introduced an activated MEK1 (MEK1 K57N) allele into the PC9 GR or H1975 cells (Fig. S7) (19). This led to WZ4002 resistance and in the resistant cells WZ4002 treatment no longer resulted in complete inhibition of ERK 1/2 phosphorylation or induction of apoptosis (Fig. S7A and S7B). In addition, MEK1 K57N was sufficient to cause resistance to both WZ4002 (data not shown) and to gefitinib (Fig. S7C) when introduced into the drug sensitive PC9 (Del E746_A750) cells. Collectively our findings suggest that activation of MAPK signaling causes WZ4002 resistance.
We further evaluated how MAPK1 amplification may prevent WZ4002 mediated apoptosis. Prior studies have demonstrated that upregulation of the pro-apoptotic protein BIM was necessary for EGFR mediated apoptosis in EGFR mutant cancers (20–22). BIM upregulation is mediated by ERK signaling (20–22). In the PC9 GR4 cells, WZ4002 treatment led to a dose dependent upregulation of BIM (Fig. 2G). In contrast, in the PC9 WZR cells, BIM upregulation was blunted consistent with the inability for WZ4002 to fully downregulate ERK 1/2 phosphorylation in these cells (Figs. 1C and 2G).
H1975 WZ4002 resistant cells retain ERK 1/2 signaling but do not contain an amplification of MAPK1
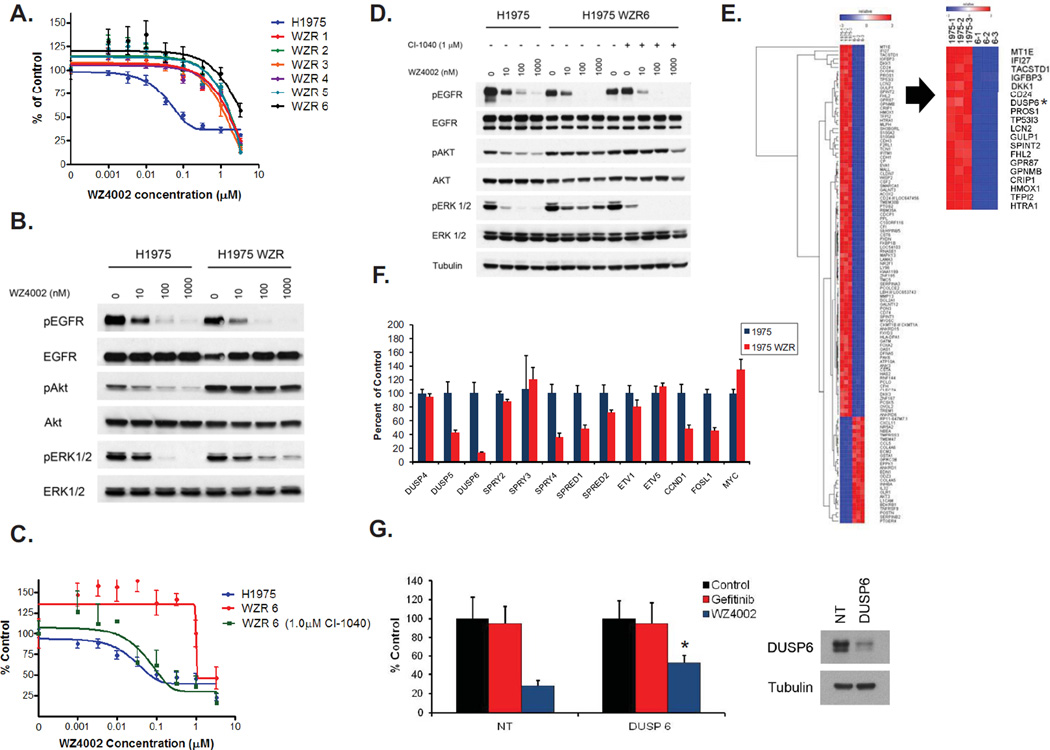
We also generated resistant versions of the WZ4002 senstitive H1975 (EGFR L858R/T790M) cell line (Fig. 3A). Similar to the PC9 WZR cells, WZ4002 was still able to inhibit EGFR phosphorylation in the H1975 WZR cells but ERK 1/2 phosphorylation was not completely inhibited (Fig. 3B) The H1975 WZR cells did not contain additional EGFR mutations, did not contain an amplification or overexpression of MAPK1 (Fig. S2B) nor harbored other regions of genomic gain when compared to the parental cells (data not shown). The MEK inhibitor CI-1040, but not the PI3K/MTOR inhibitor PI-103 (Fig. S6D), restored sensitivity to WZ4002 in the H1975 WZR cells (Fig. 3C). Furthermore, in the presence of CI-1040, WZ4002 treatment led to complete inhibition of ERK 1/2 phosphorylation as in the parental cells (Fig. 3D). In order to understand the mechanism behind sustained ERK 1/2 activation in the H1975 WZR cells we compared genome wide mRNA expression between H1975 WZR6 and H1975 cells (Fig.3E). One of the most downregulated genes in H1975 WZR cells compared to H1975 cells was DUSP6, a dual specific phosphatase that negatively regulates ERK 1/2 phosphorylation (23). We confirmed these findings using quantitative PCR and also observed downregulation of DUSP5, SPRY4 and SPRED2; all of which negatively regulate components of MAPK signaling (24–26) (Fig. 3F). We did not observe downregulation of these genes in the PC9 WZR cells (Fig. S8A). Downregulation of DUSP6 using an siRNA was sufficient to cause resistance to WZ4002 in PC9 GR4 cells (Fig. 3G) and to both gefitinib and WZ4002 in PC9 cells (Fig. S8B). Our findings suggest that downregulation of negative regulators of MAPK signaling and subsequent activation of ERK1/2 signaling is an alternative mechanism that mediates WZ4002 resistance. Furthermore, activation of ERK 1/2 signaling through introduction of MEK1 K57N into H1975 cells was also sufficient to cause WZ4002 resistance (Fig. S7D).
Figure 3.
WZ4002 resistant H1975 cells exhibit persistent ERK activation and MEK inhibition restores WZ4002 sensitivity. A. WZ4002 resistant H1975 clones. Cells were treated with WZ4002 at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. B. WZ4002 treatment does not fully inhibit ERK1/2 phosphoryaltion in WZR6 cells. H1975 and H1975 WZR6 cells were treated with increasing concentrations of WZ4002. Cell extracts were immunoblotted to detect the indicated proteins. C. CI-1040 (1 µM) restores sensitivity to WZ4002 in the WZR6 cells. D. CI-1040 (1 µM) restores the ability of WZ4002 to fully inhibit ERK 1/2 phosphorylation in the WZR6 cells. E. Comparison of expression profiles of H1975 and WZR 6 cells (left panel). Hierarchical clustering of the differentially expressed genes (p<0.0025, fold change (FC) >3.9) was performed using GENE-E. DUSP6 (asterix) ranks among the top differentially expressed genes (right panel). F. Quantitative PCR of genes in MEK/ERK transcriptional output in H1975 and H1975 WZR cells. The data are normalized to the H1975 cells. Error bars denote standard deviation. G. PC9 GR4 cells were treated with gefitinib (1 µM) or WZ4002 (100 nM) following transfection with control (NT) or DUSP6 siRNA and viable cells were measured after 48 hours of treatment and plotted (mean +/− SD) relative to untreated controls. *; p < 0.05 DUSP6 vs. NT
ERK 1/2 signaling mediates resistance to WZ4002 in a murine model of EGFR T790M lung cancer
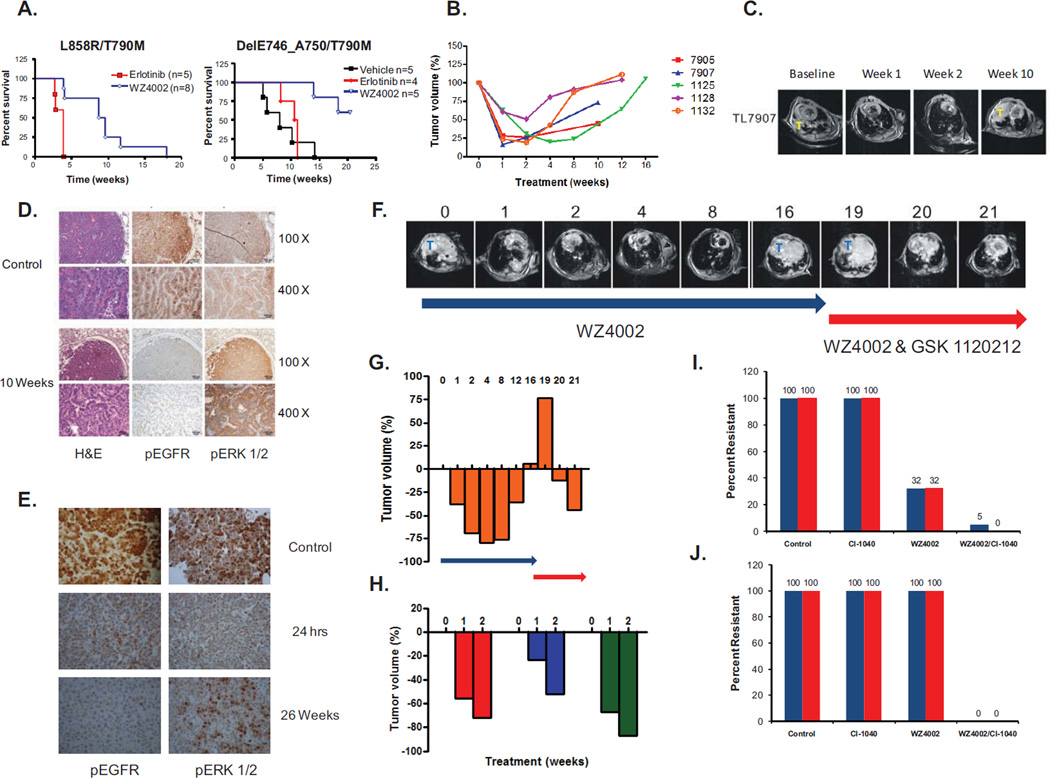
We previously demonstrated that WZ4002 is effective in vivo using murine models of EGFR T790M (L858R/T790M and DelE746_A750/T790M) NSCLC over a 2 week treatment course (13). With prolonged treatment, although we observed increased survival compared to erlotinib in both EGFR T790M bearing models (Fig. 4A), we also observed the development of acquired resistance (Figs. 4B, 4C and S9A–B). At the time of resistance we examined the tumors from the treated mice and noted that while EGFR phosphorylation was still inhibited by WZ4002, we were able detect the emergence of robust expression of ERK 1/2 phosphorylation (Figs. 4D, 4E and S10). In contrast, short term (24 hour) treatment with WZ4002 effectively inhibits both EGFR and ERK 1/2 phosphorylation in the mouse tumors (Fig 4E). We did not detect evidence of Mapk1 amplification by FISH in the resistant tumors (Fig. S11A), evidence of Kras mutations (data not shown), or loss of NF1, a negative regulator of MAPK signaling, at either the protein (Fig. S11B) or RNA (Fig. S11C) level (27). However, some of the resistant tumors had evidence of decreased Dusp6 expression compared to their drug sensitive counterparts (Fig. S12). Given the persistent ERK 1/2 signaling in the WZ4002 resistant tumors, we investigated whether a clinical MEK inhibitor, GSK 1120212, could restore the sensitivity to WZ4002 in vivo. Following the development of acquired resistance to WZ4002 (after 19, 20 and 28 weeks of therapy) we switched treatment to the combination of WZ4002 and GSK 1120212 (Fig. 4F). In 3/3 mice, GSK 1120212 restored sensitivity to WZ4002 (Figs. 4G and 4H).
Figure 4.
Development of in vivo resistance to WZ4002 in genetically engineered mouse models of EGFR T790M. A. Kaplan Meier survival curves of L858R/T790M or DelE746_A750/T790M mice treated with erlotinib, vehicle or WZ4002. Treatment with WZ4002 significantly prolongs survival compared to erlotinib (p = 0.0015 (L858R/T790M) and p = 0.0064 (Del E746_A750/T790M); both log-rank test). B. Change in tumor volume over time in EGFR L858R/T790M mice (n = 5) treated with WZ4002. Each curve represents and individual mouse. C. MRI images from mouse 7907 from B. T = Tumor. D. Immunohistochemical analyses using indicated antibodies of tumors from an EGFR L858R/T790M untreated and WZ4002 treated (10 weeks) mice. EGFR phosphorylation but not ERK 1/2 phosphorylation is inhibited. H&E; hematoxylin and eosin E. Immunohistochemical analyses using indicated antibodies of tumors from EGFR Del E746_A750/T790M untreated and WZ4002 treated (24 hours and 26 weeks) mice. WZ4002 treatment inhibits both EGFR and ERK 1/2 phosphorylation at 24 hrs but ERK 1/2 phosphorylation returns following 26 weeks of therapy. F. Serial MRI images of an EGFR L858R/T790M mouse treated with WZ4002 alone or with the combination of WZ4002 and GSK1120212. T = Tumor. G. Tumor volume measurements from mouse in F. Blue arrow, treatment with WZ4002; red arrow; treatment with both WZ4002 and GSK1120212. Numbers indicate weeks of treatment. H. Waterfall plot of tumor volume from 3 L858R/T790M mice following treatment with WZ4002 and GSK1120212. I. Long term treatment of PC9 GR4 cells with CI-1040 alone, WZ4002 alone or the combination of both drugs. The percent of resistant colonies relative to control are plotted from 2 independent experiments. J. Long term treatment of H1975 cells with CI-1040 alone, WZ4002 alone or the combination of both drugs. The percent of resistant colonies relative to control are plotted from 2 independent experiments.
An alternative strategy to treating drug resistance is to delay or prevent it from occurring. To determine whether this strategy may be applicable to the current model we evaluated this using an in vitro model. We exposed the WZ4002 sensitive PC9 GR (Fig. 4I) or H1975 (Fig 4J) cells to either WZ4002 alone, CI-1040 alone or the combination of both agents for 3 months and quantified resistant clones. CI-1040 was ineffective in both models while WZ4002 alone led to decreased number of resistant clones only in the PC9GR4 cells (Fig 4I). However, the combination of WZ4002/CI-1040 was significantly more effective at preventing the emergence of drug resistant clones in both models (Figs. 4I and 4J).
MAPK1 amplification is present in erlotinib resistant NSCLC patient
Our preclinical studies suggest that reactivation of ERK 1/2 signaling not only can mediate resistance to WZ4002 but also to geftinib in drug sensitive EGFR mutant NSCLC cell lines (Fig. S7C). We thus evaluated tumor specimens from erlotinib treated NSCLC patients that had developed drug resistance (Table S1) for presence of MAPK1 amplification. In 1 of 21 patients (4.8%) examined, we identified MAPK1 amplification which was not present in the pre-treatment drug sensitive tumor specimen (Fig. 5). The resistant tumor also lacked the more common drug resistance mechanisms EGFR T790M and MET amplification (data not shown).
Figure 5.
MAPK1 amplification is present in erlotinib resistant EGFR mutant NSCLC. FISH analysis of a baseline (left) and a post-erlotinib treated tumor (right). There is evidence of MAPK1 amplification (arrows) in the relapsed tumor. MAPK1 (red), 22q13.33 reference probe (green; RP11-768L22). Scale bar; 5 µm
MAPK1 amplification leads to an increase in EGFR internalization
We initially noticed that despite the absence of EGFR secondary mutations in the PC9 WZR cells, 10 times greater concentrations of WZ4002 was required to inhibit EGFR phosphorylation (Fig. 1C and 6A). This observation could be due to presence of mutant or wild type phosphorylated EGFR (the PC9 GR and WZR cells are both heterozygous for Del E746/A750 (data not shown)). We immunoprecipitated the mutant EGFR from the PC9 GR4 and WZR cells using a mutant specific (EGFR del E746_A750M) EGFR antibody (Fig. 6A) following WZ4002 treatment. Compared to the PC9 GR4 cells, greater concentrations of WZ4002 were required to inhibit Del E746_A750/T790M consistent with the observations in whole cell lysates (Fig. 6A). One potential explanation for these differences is an alteration in EGFR internalization. An alteration in EGFR internalization could lead to changes in cellular localization of EGFR which may impact drug access and hence the need for an increased drug concentration required to inhibit EGFR phosphorylation. We evaluated EGFR internalization using 125I labeled EGF. In both the WZR10 and 12 cells, we observed a significantly larger ke than in the GR4 cells (Fig. 6B). There was no difference between the PC9 GR4 and the PC9 cells (Fig. 6B). Inhibition of MEK using CI-1040 did not alter ke in GR4 cells, but significantly, although not completely, reduced EGFR phosphorylation and ke in the WZR cells both to levels similar to that observed in the parental PC9 GR4 cells (Figs. 6C and 6D). Prior studies have implicated Thr 669 of EGFR as an ERK phosphorylation site and associated with EGFR turnover possibly through internalization (28–30). This site is highly phosphorylated in the PC9 WZR cells compared to the parental PC9 GR4 cells (Fig. 6E) and is inhibited by CI-1040 treatment alone without effecting the EGFR autophosphorylation site on Tyr 1068 (Fig. 6E). We did not observed any changes in ke in the H1975WZR cells (data not shown).
Figure 6.
MAPK1 amplification alters EGFR internalization. A. PC9 GR4 or WZR10 cells are treated with increasing concentrations of WZ4002 for 6 hrs. Cell extracts from whole lysates (top) or following immunopercipitation with an EGFR DelE746_A750 antibody (bottom) were immunoblotted to detect the indicated proteins. B. Internalization rate constants (ke) for 125I labeled EGF in different cell lines. The ke is significantly greater for the WZR cells compared to the PC9 GR4 cells. C. PC9 GR4 and WZR 10 cells treated with WZ4002 alone or in combination with CI-1040 for 6 hours. Cell extracts following immunopercipitation with an EGFR DelE746_A750 antibody were immunoblotted to detect the indicated proteins. D. Internalization rate constants (ke) for 125I labeled EGF in following treatment with CI-1040 (3 mM) for 24 hrs. There is a significant reduction in ke in the WZR10 cells with CI-1040 treatment. E. EGFR phosphorylation at Thr-669 is markedly increased in WZR10 compared to GR4 cells. CI-1040 alone inhibits phosphorylation at Thr-669 but not Tyr-1068.
Successive tyrosine kinase inhibitor resistance also leads to chemotherapy resistance
Following the development of clinical resistance to EGFR kinase inhibitors NSCLC patients are often treated with systemic chemotherapy. Our successive cell line models (PC9; gefitinib and WZ4002 sensitive, PC9 GR; gefitinib resistant but WZ4002 sensitive and PC9 WZR; gefitinib and WZ4002 resistant) provides a system in which to evaluate whether kinase inhibitor resistance impacts chemotherapy sensitivity. We treated the cells with staurosporine, paclitaxel and etoposide. As can be seen in Figure 7A, these agents induce the greatest degree of cell death in the parental PC9 cells and the least in the PC9WZR cells. All three of these agents, as well as EGFR inhibitors, are known to kill via the mitochondrial pathway of apoptosis, suggesting that cells may select for resistance to apoptosis more generally when they select for resistance to tyrosine kinase inhibition (21). MEK inhibition in PC9 WZR cells did not reverse resistance to chemotherapy (data not shown). It has recently been shown that one can measure pre-treatment proximity of the mitochondria to the apoptotic threshold, known as mitochondrial priming, using BH3 profiling (31). Proximity to the threshold, indicated by increased mitochondrial depolarization in response to BH3 peptides, was shown to correlate to chemosensitivity across a broad range of cancers (31). We found that the successive generations of kinase inhibitor resistant PC9 cells were indeed less primed (Fig. 7B). This result suggests that a more broad resistance to apoptotic signaling may be selected for when cells select for TKI resistance.
Figure 7.
EGFR TKI resistant PC9 cells are also more resistant to cytotoxic chemotherapy. A. Cell viability experiments following 24 h exposure to staurosporine (1 µM), paclitaxel (1 µM) or etoposide (100 µM). DMSO is used as a control. The mean values and standard deviation from 3 independent experiments is shown. B. Mitochondrial depolarization response to BH3 peptides for PC9, PC9 GR4 and WZR10 cells. The mean values and standard deviation from 3 independent experiments are shown.
Discussion
Despite the dramatic clinical efficacy of EGFR kinase inhibitors in EGFR mutant NSCLC, single agent EGFR inhibitors will not cure advanced NSCLC. Studies of drug resistance mechanisms provide insights into how cancers develop resistance and the findings from these studies can be used to design rational combination therapeutic strategies (14). EGFR T790M is the most common mechanism of acquired drug resistance to erlotinib and gefitinib and, to date, has also been the most difficult to treat clinically (5–7, 11). This is mirrored by our prior preclinical studies using a clinical irreversible quinazoline EGFR inhibitor (PF299804 or dacomitinib) (12). Although the PC9 GR cells are transiently sensitive to PF299804, resistant cells harboring EGFR T790M amplification rapidly emerge (12). These EGFR T790M amplified cells retain sensitivity to the mutant selective EGFR inhibitor WZ4002 (Fig. 1A) and remarkably EGFR T790M amplification never emerges as a resistance mechanism when PC9 GR4 cells are exposed to WZ4002. These findings not only highlight the potential clinical efficacy of a more potent EGFR inhibitor but also the rapid ability of cancer cells to adapt to such an inhibitor as we are still able to select WZ4002 resistant clones from the PC9 GR cells.
Previous studies have implicated maintenance of PI3K signaling as critical in mediating resistance to EGFR kinase inhibitors (32, 33). Resistance mechanisms to EGFR TKIs include MET amplification, HGF production and PIK3CA mutations, all of which block gefitinib or erlotinib mediated inhibition of AKT phosphorylation (14, 32, 34). Inhibition of PI3K signaling using PI-103 alone or in combination with gefitinib has been demonstrated to overcome HGF mediated resistance to gefitinib both in vitro and in vivo (35). Our current findings suggest that resistance to EGFR inhibitors can also arise through persistent or re-activation of ERK1/2 signaling. This can occur through at least two independent mechanisms: genomic amplification of MAPK1 and downregulation of negative regulators of ERK1/2 signaling (Figs. 1D and 3F). These drug resistant cells are not ERK dependent as MEK or ERK inhibition alone is not sufficient to restore apoptosis but rather requires concomitant EGFR inhibition (Figs. 2B and 2D). In contrast to HGF mediated resistance, PI3K or AKT inhibition alone or in combination with WZ4002 is not sufficient to reverse drug resistance since PI3K inhibition does not result in ERK 1/2 inhibition (Figs. S6) (35). Thus the therapeutic strategy for overcoming EGFR inhibitor resistance needs to be tailored based on the specific signaling pathways activated by each of the resistance mechanisms.
We further demonstrate that our findings have clinical relevance as MAPK1 amplification can also emerge in erlotinib resistant EGFR mutant NSCLC patients (Fig. 5). The frequency at which this occurs is low (Table S1) but not unexpected given the high prevalence of EGFR T790M as an erlotinib resistance mechanism (7). This observations may be due to the pre-existence of EGFR T790M in some treatment naïve cancers coupled with the current lack of effective clinical therapies against EGFR T790M (11, 36). This hypothesis is supported by preclinical studies of the PC9 cells where multiple studies demonstrate the emergence of EGFR T790M following exposure to first or second generation EGFR TKIs (37–39). In contrast, WZ4002 resistant PC9 cells do not harbor EGFR T790M (13). As EGFR T790M directed inhibitors, including CO-1686 (NCT01526928), enter clinical development, MAPK1 amplification may begin to emerge as a more common resistance mechanism, and should be evaluated, along with other mechanisms leading to reactivation of ERK signaling, in tumor specimens when clinical drug resistance develops.
Our study also identifies two unique aspects of drug resistance mediated by MAPK1 amplification and serve to highlight the complexity of drug resistance mechanisms. In addition to its effects on signaling, MAPK1 amplification correlates with changes in EGFR internalization (Fig. 6B). This leads to a 10 fold increase in the concentration of WZ4002 required to fully inhibit EGFR phosphorylation (Fig. 6A). Although this difference may at first glance seem subtle, in cancer patients receiving therapy with a kinase inhibitor, this change in drug concentration required for target inhibition could be the difference between a clinically sensitive and resistant tumor. Previous studies have demonstrated a reduced rate of ligand mediated EGFR internalization in drug sensitive EGFR mutant NSCLC cell lines (40). The decreased rate of EGFR endocytosis was found to be associated with an impaired ability of EGFR to fully utilize SHP2 for complete activation of ERK signaling (40). In contrast, enhanced ERK signaling, as observed in the PC9 WZR cells, plays a causal role in increased EGFR endocytosis. This observation may be due to the ability of ERK to phosphorylate EGFR at Thr-669, as observed in the WZR cells (Fig. 6E), leading to altered EGFR trafficking (28–30). Although clinically relevant, it will remain a challenge to study changes in receptor internalization from clinical diagnostic specimens unless they are due to a genomic alteration as in the current study. Our findings also reveal that sequential resistance to kinase inhibitors (gefitinib and WZ4002) renders EGFR mutant NSCLC cells less susceptible to chemotherapeutic agents (Fig. 7B). These observations are potentially clinically significant as EGFR kinase inhibitors are currently being used as initial therapy for EGFR mutant NSCLC and may ultimately impact the sensitivity to a broad range of subsequent therapies (2, 3). These findings also serve to highlight that drug resistance may not simply be an alteration in one signaling pathway but rather a more complex process that more broadly impacts apoptotic signaling.
Findings from the our study demonstrate that the combination of WZ4002 and an allosteric MEK inhibitor may be an effective strategy not only to treat drug resistant cancers (Figs. 2B, 3C, 4G and 4H) but also to prevent the emergence of drug resistant clones (Fig. 4I and 4J). Coupled with our prior studies, demonstrating that WZ4002 alone can prevent the emergence of EGFR T790M in model systems, the combination of WZ4002 and a MEK inhibitor may be a particularly effective therapeutic strategy for EGFR mutant NSCLC and should be tested in a clinical trial (13).
Methods
Cell Culture and reagents
The EGFR mutant NSCLC cell lines PC9 (delE746_A750), PC9 GR4 (delE746_A750/T790M), and H1975 (L858R/T790M) have been previously characterized (12, 13, 41). The PC9 and H1975 cells were confirmed by fingerprinting using the Power Plex 1.2 system (Promega, Madison, WI) most recently in March 2012. Gefitinib, CI-1040 and MK-2206 were obtained from Selleck chemicals (Houston, TX). WZ4002 was synthesized using previously published methods (13). GSK1120212 was synthesized at Haoyuan Chemexpress (Shanghai, China) according to published methods (42). Compound 11e was synthesized as in (15). Stock solutions of all drugs were prepared in DMSO and stored at −20°C. DUSP6 and control (non-targeting; NT) siRNA reagents were obtained from Dharmacon (Lafayette, CO) and used according to the manufacturer’s recommended conditions.
Cell proliferation and growth assays
Growth and inhibition of growth was assessed by MTS assay according to previously established methods (32, 43, 44). All experimental points were set up in six to twelve wells and all experiments were repeated at least three times. The data was graphically displayed using GraphPad Prism version 5.0 for Windows, (GraphPad Software Inc., La Jolla, CA). Long term drug treatment studies were performed in 96 well format using 100 wells for each condition and performed in triplicate. Resistant wells were assayed following 3 months of treatment and are plotted relative to untreated cells.
Antibodies and Western Blotting
Cells grown under the previously specified conditions were lysed in NP-40 buffer. Western blot analyses were conducted after separation by SDS/PAGE electrophoresis and transfer to PVDF-P immobilon membranes (Millipore). Immunoblotting was performed according to the antibody manufacturers' recommendations. Anti-phospho-Akt (Ser-473), anti-total-Akt, EGFR delE746_A750 specific, phospho (Thr669) EGFR, pRSK, total-RSK, DUSP6, and BIM antibodies were obtained from Cell Signaling Technology. The phospho-EGFR (pY1068), total-ERK1/2, phospho-ERK1/2 (pT185/pY187) antibodies were purchased from Biosource International Inc. Total EGFR antibody was purchased from Bethyl Laboratories. The NF1 antibody was used as previously described (45).
Generation of drug resistant cell lines
To generate drug resistant cell lines, NSCLC cells were exposed to increasing concentrations of WZ4002 similar to previously described methods (12, 14, 32). Individual clones from WZ4002 resistant (WZR) cells were isolated and confirmed to be drug resistant.
EGFR mutational analyses
Total RNA was isolated from cell lines or tumors using Trizol™ (Invitrogen, Carlsbad, CA) and purified using RNeasyTM minielute cleanup kit (Qiagen,Valencia, CA). cDNA was transcribed with Superscript II Reverse Transcriptase (Invitrogen Life technologies, Carlsbad, CA) and used as template for subsequent PCR based studies (14, 32). The PCR products were also cloned into a TOPO TA vector (Invitrogen, Carlsbad, CA), transformed into bacteria and the inserts from individual clones sequenced. The PCR primers and conditions are available upon request.
SNP analyses
SNP analyses were performed as previously described (14). Samples were processed for the Human Mapping 250K Sty single nucleotide polymorphism (SNP) array according to the manufacturer's instructions. Comparison of gene copy number differences was performed using the dChip software according to previously established methods (14, 46). The SNP data have been deposited to GEO (accession number GSE37700).
MAPK1 copy number analysis
The absolute copy number for MAPK1 was determined using quantitative real time PCR as previously described (14). Quantification was based on standard curves from a serial dilution of normal human genomic DNA. All specimens were analyzed in triplicate. The PCR primers are available upon request.
FISH probes and hybridization
Human MAPK1 probe consisted of a mixture of three fosmids (WI2-1114H9, WI2-1468D5 and WI2-3011D5) and spanned the entire gene locus. A bacterial artificial chromosome (BAC) (RP11-768L22) located on 22q13.33 was used as a reference probe. A BAC (RP24-174O22) was used as the murine Mapk1 probe and BAC RP23-122A24 located on chromosome 16QA1 was used as a reference probe. All fosmids and BACS were purchased from Children’s Hospital Oakland Research Institute (CHORI; Oakland, CA). DNA was extracted using a Qiagen kit (Qiagen Inc., Valencia CA) and labeled with Spectrum Green- or Spectrum Orange-conjugated dUTP by nick translation (Vysis/Abbott Molecular, Des Plaines, IL). The CEP7 probe (Vysis/Abbott Molecular, Des Plaines, Il) was used according to the manufacturer’s instructions. Chromosomal mapping and hybridization efficiency for each probe set were verified in normal metaphase spreads (data not shown). Three color FISH assays were performed as previously described (12, 14). Tumor cells were classified as containing a MAPK1 amplification if the ratio of MAPK1/reference ≥ 2 or if there were ≤ 1 reference signals and ≥ 3 MAPK1 signals.
Generation of Mouse Cohorts and Treatment with WZ4002
Doxycycline inducible EGFR-TL (L858R/T790M) and EGFR-TD (DelE746_A750/T790M) transgenic mice were generated as previously described (13, 47). All mice were housed in a pathogen- free environment at the Harvard School of Public Health and were handled in strict accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare, and all animal work was done with Dana-Farber Cancer Institute IACUC approval.
Cohorts of EGFR TL/CCSP-rtTA and EGFR TD/CCSP-rtTA were put on doxycycline diet at 6 weeks of age to induce the expression of mutant EGFR. Mice were evaluated by MRI after 12 to 16 weeks of doxycycline diet to document and quantify the lung cancer burden before being assigned to various treatment study cohorts. Tumor bearing mice were treated either with vehicle (NMP (10% 1-methyl-2-pyrrolidinone: 90% PEG-300) alone or WZ4002 at 50 mg/kg gavage daily. Following the development of WZ4002 resistance, mice were treated with both WZ4002 (50 mg/kg daily oral gavage) and GSK 1120212 (1.5 mg/Kg daily by oral gavage). MRI evaluation was repeated every 2 weeks. MRI and tumor burden measurement were performed as described previously (47, 48).
MRI Scanning and Tumor Volume Measurement
Mice were anesthetized with 1% isoflurane in an oxygen/air mixture. The respiratory and cardiac rates of anesthetized mice were monitored using Biotrig Software. The animals were imaged with a rapid acquisition with relaxation enhancement (RARE) sequence (TR = 2000 ms, TE effect = 25 ms) in the coronal and axial planes with a 1mm slice thickness and with sufficient number of slices to cover the entire lung. Matrix size of 128 × 128 and a field of view (FOV) of 2.5 cm × 2.5 cm2 were used for all imaging. With same geometry and described above, the mice were also imaged with a gradient echo fast imaging (GEFI) sequence (TR = 180 ms, TE effect = 2.2 ms) with respiratory and cardiac gating, in both coronal and axial planes. The detailed procedure for MRI scanning has been previously described (47, 48). The tumor burden volume and quantification were reconstructed using 3D slicer software (49).
Immunohistochemical Analyses
Hematoxylin and eosin (H&E) staining of tumor sections was performed at the Department of Pathology at the Brigham and Women’s Hospital. Immunohistochemistry for pEGFR and pERK 1/2 was performed on formal fixed paraffin embedded tumor sections using previously described methods (13).
EGF radiolabeling and measurement of EGF internalization rate constants
Recombinant human EGF (Peprotech, Rocky Hill, NJ, USA) was labeled with 125I (PerkinElmer, Waltham, MA, USA) in the presence of an iodobead catalyst (Thermo Scientific, Waltham, MA, USA), as described previously (50). The activity of the labeled EGF was determined using a phosphotungstic acid precipitation assay. To measure rate constants for labeled EGF internalization (ke), serum-starved cells were exposed to 10 ng/mL 125I-EGF at 37°C for up to 7.5 min. At five evenly spaced time points, cells were quickly washed with a buffer to remove bulk ligand, incubated in a mild acid strip solution to obtain surface-associated ligand, and finally solubilized in 1 N NaOH to obtain internalized ligand. Buffer washes and incubations were done at 4°C to minimize further EGFR internalization during these steps. 125I-EGF counts in surface and internal fractions were quantified using a 1470 Wizard Gamma Counter (PerkinElmer). With these data, ke values were calculated using a simple kinetic model of ligand-mediated receptor internalization, as described previously (50). Measurements were corrected for 125I-EGF spillover from acid stripping and non-specific binding of 125I-EGF to the cell surface. For some measurements, cells were pretreated for ~24 hrs 3 µM CI-1040.
RNA expression profiling and quantitative PCR
Total RNA was prepared from drug sensitive and resistant cells as described above. Synthesis of cRNA and hybridization to Human Expression Array U133A2.0 chips were performed following Affymetrix protocols (Affymetrix, Inc.). Probe-level intensity data files in the CEL format were pre-processed using Robust Multichip Average program using the Gene Pattern software (Broad Institute, Cambridge, MA). Probes representing the same genes were collapsed into a single value, and standardized by taking the median value for each gene across sample set in the GenePattern software. Comparative maker selection module was used to select differentially expressed genes that meet defined criteria (p<0.0025, fold change (FC) >3.9). Hierarchical clustering of the differentially expressed genes that meets the criteria was performed using GENE-E (Broad Institute, Cambridge, MA). The expression data have been deposited to GEO (accession number GSE37700). Quantitative PCR to evaluate expression of genes associated with MEK/ERK-dependent transcriptional output was performed in triplicate as described in (24). NF1 expression was performed using the NF1-TaqMan Gene Expression Assay (Mm00812424_m1; Applied Biosystems).
BH3 profiling
BH3 profiling was performed as previously described (31).
FACS analyses
Cell viability experiments were performed using drug sensitive and resistant cell lines exposed following drug exposure for 24 to 72 hrs. Cells were stained with fluorescent conjugates of annexin-V (BioVision) and/or propidium iodide (PI) and analyzed on a FACSCanto machine (Becton Dickinson). Viable cells are annexin-V- and PI-, and cell death is expressed are 100%-viable cells.
Patients
NSCLC patients treated with erlotinib were identified from the Thoracic Oncology Program at DFCI. Tumor biopsies at the time of relapse were obtained under an IRB approved protocol. Analyses for EGFR T790M and MET amplification were performed as previously described (14). All patients provided written informed consent.
Supplementary Material
Significance.
Our study identifies activated ERK signaling as a mediator of resistance to irreversible pyrimidine EGFR inhibitors in EGFR T790M bearing cancers. We further provide a therapeutic strategy to both treat and prevent the emergence of this resistance mechanism.
Acknowledgements
This study is supported by grants from the National Institutes of Health RO1CA114465 (P.A.J.), R01CA135257 (P.A.J.), PO1CA154303 (P.A.J., K.-K.W. and N.S.G.), K08CA127350 (C.A.P.), National Cancer Institute Lung SPORE P50CA090578 (P.A.J. and K.-K.W.), the Cammarata Family Foundation Research Fund (M.C. and P.A.J.), the Nirenberg Fellowship at Dana-Farber Cancer Institute (M.C. and P.A.J.) and the by grant #IRG-78-002-30 from the American Cancer Society (M.J.L. and C.S.M.).
Footnotes
Potential conflicts of interest for all authors. All authors have completed separate conflict of interest forms.
Dalia Ercan, Chunxiao Xu, Masahiko Yanagita, Calixte S. Monast, Christine A. Pratilas, Joan Montero, Mohit Butaney, Takeshi Shimamura, Lynette Sholl, Elena Ivanov, Madhavi Tadi, Andrew Rogers, Claire Repellin, Marzia Capelletti, Ophélia Maertens, Eva M. Goetz, Matthew J. Lazzara, Neal Rosen, Kwok-Kin Wong
No conflicts of interest
References
- 1.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012 doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 3.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005;2:1–11. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008 doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 11.Miller VA, Hirsh V, Cadranel J, Chen Y-M, Park K, Kim S-W, et al. Phase IIB/III Double-Blind Randomized Trial of Afatinib (BIBW2992, an Irreversible Inhibitor of EGFR/HER1 and HER2) + Best Supportive Care (BSC) Versus Placebo + BSC in Patients with NSCLC Failing 1–2 Lines of Chemotherapy and Erlotinib or Gefitinib (LUX-LUNG 1) Annals of Oncology. 2010;21:LBA1. [Google Scholar]
- 12.Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutantselective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 15.Aronov AM, Tang Q, Martinez-Botella G, Bemis GW, Cao J, Chen G, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J Med Chem. 2009;52:6362–6368. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 16.Dalby KN, Morrice N, Caudwell FB, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 17.Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–5850. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 18.Tan S, Ng Y, James DE. Next-generation Akt inhibitors provide greater specificity: effects on glucose metabolism in adipocytes. Biochem J. 2011;435:539–544. doi: 10.1042/BJ20110040. [DOI] [PubMed] [Google Scholar]
- 19.Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. discussion 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–11875. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- 22.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFRdependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- 24.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dualspecificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 26.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 27.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Huang Y, Jiang J, Frank SJ. ERK-dependent threonine phosphorylation of EGF receptor modulates receptor downregulation and signaling. Cell Signal. 2008;20:2145–2155. doi: 10.1016/j.cellsig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takishima K, Griswold-Prenner I, Ingebritsen T, Rosner MR. Epidermal growth factor (EGF) receptor T669 peptide kinase from 3T3-L1 cells is an EGF-stimulated "MAP" kinase. Proc Natl Acad Sci U S A. 1991;88:2520–2524. doi: 10.1073/pnas.88.6.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Northwood IC, Gonzalez FA, Wartmann M, Raden DL, Davis RJ. Isolation and characterization of two growth factor-stimulated protein kinases that phosphorylate the epidermal growth factor receptor at threonine 669. J Biol Chem. 1991;266:15266–15276. [PubMed] [Google Scholar]
- 31.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donev IS, Wang W, Yamada T, Li Q, Takeuchi S, Matsumoto K, et al. Transient PI3K inhibition induces apoptosis and overcomes HGF-mediated resistance to EGFRTKIs in EGFR mutant lung cancer. Clin Cancer Res. 2011;17:2260–2269. doi: 10.1158/1078-0432.CCR-10-1993. [DOI] [PubMed] [Google Scholar]
- 36.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 38.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, et al. Optimization of Dosing for EGFR-Mutant Non-Small Cell Lung Cancer with Evolutionary Cancer Modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, et al. The T790M "gatekeeper" mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 40.Lazzara MJ, Lane K, Chan R, Jasper PJ, Yaffe MB, Sorger PK, et al. Impaired SHP2-mediated extracellular signal-regulated kinase activation contributes to gefitinib sensitivity of lung cancer cells with epidermal growth factor receptor-activating mutations. Cancer Res. 2010;70:3843–3850. doi: 10.1158/0008-5472.CAN-09-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signalregulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–472. [PubMed] [Google Scholar]
- 42.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 43.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 44.Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97:1185–1194. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- 45.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 47.Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 49. www.slicer.org.
- 50.Lund KA, Opresko LK, Starbuck C, Walsh BJ, Wiley HS. Quantitative analysis of the endocytic system involved in hormone-induced receptor internalization. J Biol Chem. 1990;265:15713–15723. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.