Summary
Different trans-acting factors (TF) collaborate and act in concert at distinct loci to perform accurate regulation of their target genes. To date, the co-binding of TF pairs has been investigated in a limited context both in terms of the number of factors within a cell type and across cell types and the extent of combinatorial co-localizations. Here we use a novel approach to analyze TF co-localization within a cell type and across multiple cell lines at an unprecedented level. We extend this approach with large-scale mass spectrometry analysis of immunoprecipitations of 50 TFs. Our combined approach reveals large numbers of interesting and novel TF-TF associations. We observe extensive change in TF co-localizations both within a cell type exposed to different conditions and across multiple cell types. We show distinct functional annotations and properties of different TF co-binding patterns and provide new insights into the complex regulatory landscape of the cell.
Introduction
Trans-acting factors operate cooperatively to regulate gene expression across various cell types and environmental conditions. Previous studies have shown that different factors bind in concert at cis-regulatory modules and either collaborate or compete to achieve complex and accurate regulation of target genes. Systematic assays of TF co-binding have been performed and analyzed in lower organisms, such as E. coli (Balázsi et al., 2005), yeast (Lee et al., 2002), and the Drosophila embryo (Lifanov et al., 2003; Segal et al., 2008). However, these studies have largely been limited to computational prediction of co-localized binding or a limited number of datasets and are thus subject to a large number of false positive sites and do not necessarily represent co-localized binding in a specific cell state.
Recently, the ENCODE consortium has described ChIP-seq analysis of 125 trans-acting factors (including 119 DNA-binding factors) in 72 human cell lines (76 in K562 cells)(The ENCODE Project Consortium, 2012). These data have begun to reveal complex co-localization patterns driving regulatory function (Gerstein et al., 2012). However, these studies primarily focused on a single cell type (K562) and analyzed a limited number of factors. Moreover, TF co-localizations were primarily studied in the context of the binding region for one factor, which greatly limited the number of potential co-localizations that could be observed. Thus, a global understanding of TF binding was not evident within or across multiple cell types, nor was the co-localization investigated in an unbiased fashion. Furthermore, the dynamics of TF binding was not examined.
Here we present a novel approach using an unbiased machine learning method to investigate in detail the co-localization of TFs within a single cell type and across multiple cell types. The ChIP-seq data used contains 128 TF binding datasets in a single cell type (K562) as well as over 50 factors in multiple cell types. This is an increase of 83 TF binding datasets over the previously published ENCODE data. We find an unprecedented number of novel co-localizations and dynamic changes in TF co-localizations. We integrate these findings with protein-protein interactions identified by mass spectrometry using the same antibodies for the ChIP-seq analysis. We show the subset of co-localizations that are due to direct binding within complexes and those that are due to independent recruitment of TFs to the DNA. Overall our results provide many insights into TF co-localizations that define the regulatory code of humans.
Results
Self-organizing Map and the Overall Rationale
The study of the co-binding of TFs in large data sets is difficult due to the high dimensionality of the data. For example, exploration of the complete space of combinatorial binding for 128 TF datasets is not feasible as there are more than 1038 possible combinations of binding. Because of this, previous work explored this problem in a limited fashion using either enrichment of pairs of binding factors in a specific context (e.g. at promoter regions) (Chikina and Troyanskaya, 2012) or binding of pairs of factors in the context of a specified factor (Gerstein et al., 2012). In order to test the full combinatorial space without delineating all possible combinations, we employed an artificial neural network called a self-organizing map (SOM) which organizes the TF binding data in an unsupervised manner (Kohonen, 2001). SOMs have been successfully used in a large number of applications and have proven to be robust and accurate (Tamayo et al., 1999; The ENCODE Project Consortium, 2012). This technique is ideal for displaying the high-dimensional information of TF co-localizations while retaining topological properties of the data. This property allows for a map of the data to be projected in two-dimensions with more similar patterns of binding in closer proximity. Furthermore, once a map of TF co-binding is generated, it is possible to explore it for a very wide range of additional properties such as levels of expression and gene ontology information.
In addition to co-localized binding of individual factors at genomic loci, some of the apparent co-binding in ChIP-seq data may be due to direct protein-protein interaction (PPI) between TFs resulting in heterodimers or more elaborate complexes. Many well studied co-binding TF pairs are due to direct PPI between the TFs; for example, the well-known regulator AP-1 represents a heterodimer of members of the FOS, JUN, ATF, and JDP protein families (Hess et al., 2004). To study the relationship between TF PPI and TF co-binding in a high-throughput fashion, we performed mass spectrometry analysis on immunoprecipitation samples of 50 TFs. These mass spectrometry data together with the ChIP-Seq data enable an integrative analysis to determine TF co-localizations resulting from DNA-directed binding or PPI-directed binding.
Extensive TF Co-Binding Patterns of K562 Cells
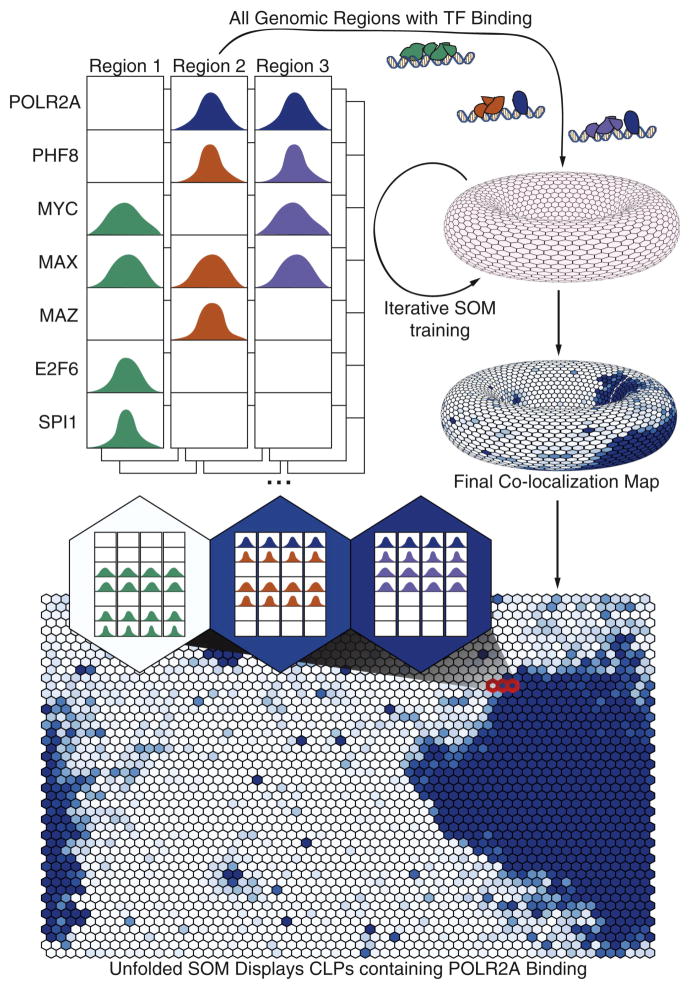
We first examined TF co-localization within a single cell type by selecting an optimally trained SOM built using 128 ChIP-seq datasets from K562 cells (See Supplemental Information for SOM generation methods, http://snyderlab.stanford.edu/SOM/). The end result is a toroidal map comprised of a series of “neurons”, each of which contains a common binding pattern found at distinct genomic locations. These neurons are depicted as flat in Figure 1 (both the left and right sides of our figures and the top and bottom parts are connected). In addition, there are no specific boundaries on the map meaning that a set of neurons in the network (represented by a hexagon) may be members of the same “cluster” of patterns. This property allows us to group neurons to identify high-level rules of factor co-localization or to zoom to a fine resolution of co-localization patterns. However, we will refer to each neuron as a distinct TF co-localization pattern (CLP) and containing a set of genomic TF binding regions called cis-regulatory modules (CRMs). Most of the TF co-localizations within a single CLP are statistically significant (See Supplemental Information, Fig. S1).
Fig. 1.
Workflow of SOM training process. At each genomic region bound by at least two TFs, the binding state of all TFs overlapping the region is encoded into binary states (binding / not binding). These binary state vectors are input into an empty SOM depicted as the gray toroid. After training each co-localization pattern (represented by a hexagon) can be represented by a specific binding pattern of TFs. Here we shade the toroid with POLR2A binding values (blue represents higher POLR2A binding in a given CLP). The SOM can then be ‘unwrapped’ for easier display of the same data. We then display that each CLP represents a group of CRMs which maintain the same binding pattern. See also Figure S1.
The SOM is able to capture the vast complexity of co-localization patterns resulting in identification of many previously known interactions. For example, we identify JUN+FOS interactions resulting in the known AP-1 complex which functions in early response to stimulus (Hess et al., 2004) (Fig. 2). The known co-binding patterns also follow known rules for their interaction. For example, CTCF+RAD21+SMC3 interact as part of the cohesin complex (Hou et al., 2010). However, SMC3 does not directly bind DNA and, as such, we find it in our SOM only in the context of CTCF and RAD21. This property also holds true for the factor NFE2 which is known to bind DNA mediated by a MAFF and MAFK heterodimer (Igarashi et al., 1995).
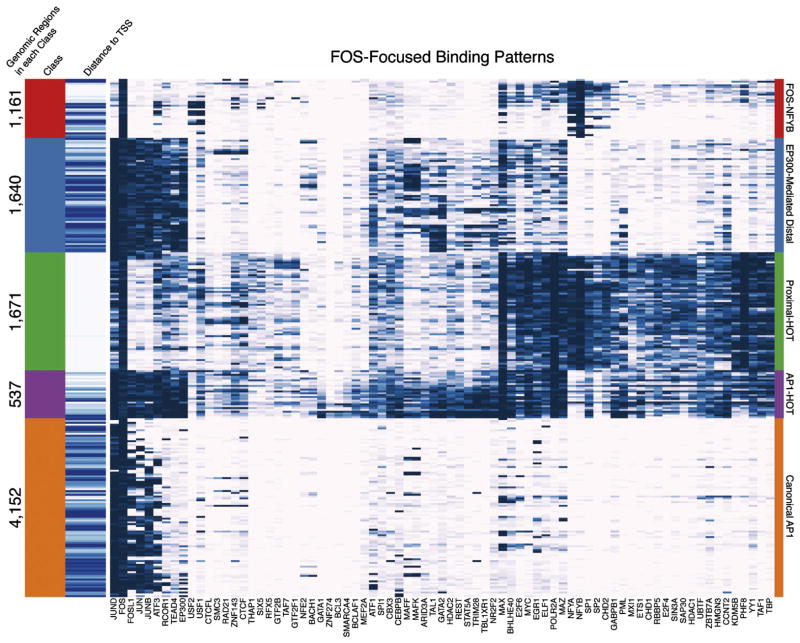
Fig. 2.
FOS-Focused binding patterns. FOS containing co-localization patterns are clustered and shown as each row of a heatmap with blue indicating signal for each co-localized factor (columns). The FOS-focused co-localization patterns fall into 5 classes: FOS-NFYB, EP300-Mediated Distal, Proximal-HOT, AP1-HOT, and Canonical AP1, which are tagged with different colors. The number of genomic regions and distance to the closest TSS (white = proximal, blue = distal) for each class of co-localization pattern is shown on the left of the heatmap. See also Figure S2.
The SOM is also able to reveal context dependent TF co-binding as reported in previous work. For example, it was shown that FOS has different partners under the contexts of proximal or distal TF binding (Gerstein et al., 2012). Our analysis recapitulates this result; however, we found that the previous interpretation is a significant oversimplification (Fig. 2). In fact, the many co-localization patterns of FOS reveal that it falls into at least 5 overarching categories: 1) “Canonical AP1” where FOS+JUND is the major co-binding along with FOSL1, JUNB, and ATF3 (other possible AP1 members) at sites distal to the transcription start site (TSS); 2) “EP300 Mediated Distal” where the canonical AP1 complex co-binds with RCOR1+TEAD4+EP300 typically at sites distal to the TSS and likely representing an enhancer state; 3) a novel “FOS+NFYB” category where FOS does not co-bind with JUND and instead has NFYA and NFYB co-binding; 4) “Proximal-HOT” where FOS+MAX+POLR2A+NFYB+PHF8 have firm co-binding along with frequent additions of many additional factors; 5) “AP1-HOT” where many TFs co-bind with the AP1 complex. We validated the novel FOS+NFYB interaction context that represents approximately 8.1% and 11.5% of all FOS and NFYB binding events respectively with a co-immunoprecipitation (Fig. S2). Furthermore, while this paper was in review an independent observation of this co-localization was reported (Fleming et al., 2013).
We find exceptional complexity in the context of some TF co-localization sets. These represent what can be considered primary binding partners that take part in a large cohort of combinatorial regulation. Several canonical, prevalent TF co-localization modules, such as CTCF+RAD21+SMC3, EP300+TAL1, and POL2+TAF1, are shown to be in the most complex contexts, as evidenced by the large number of CLPs that include these interactions. Many (but not all) of these TF co-localizations with large binding complexity are those interactions that are previously known, likely because of their high frequency of co-occurrence in the genome. We demonstrate this with a complexity network demonstrating the most frequently interacting pairs (Fig. S2). As shown in the figure, many of the canonical expected interacting TFs have frequent additional co-localizations.
In addition to confirming known interactions, we find additional co-localization patterns that have not been previously documented. These may exist as an additional factor binding in the context of known “primary” partners or as entirely novel combinations. In fact, a majority of the co-localization patterns represent more complex combinations of binding in conjunction with common themes which, given the more detailed context, are specifically enriched for different regulatory functions. For example, the CTCF+RAD21+SMC3+ZNF143 pattern is typically repressive; however, the association of JUND+MAZ with this same complex forms a novel binding pattern that up regulates its targets as evidenced by significantly higher expression of associated genes (Median expression RPKM of 0.03 and 9.11 respectively, p-value < 1×10−6, Mann-Whitney U Test). These gene targets are significantly enriched for members of the inflammasome that is a component of the innate immune system expressed in myeloid cells. For all discovered interactions and their associated annotations, see http://snyderlab.stanford.edu/SOM/.
HOT Regions
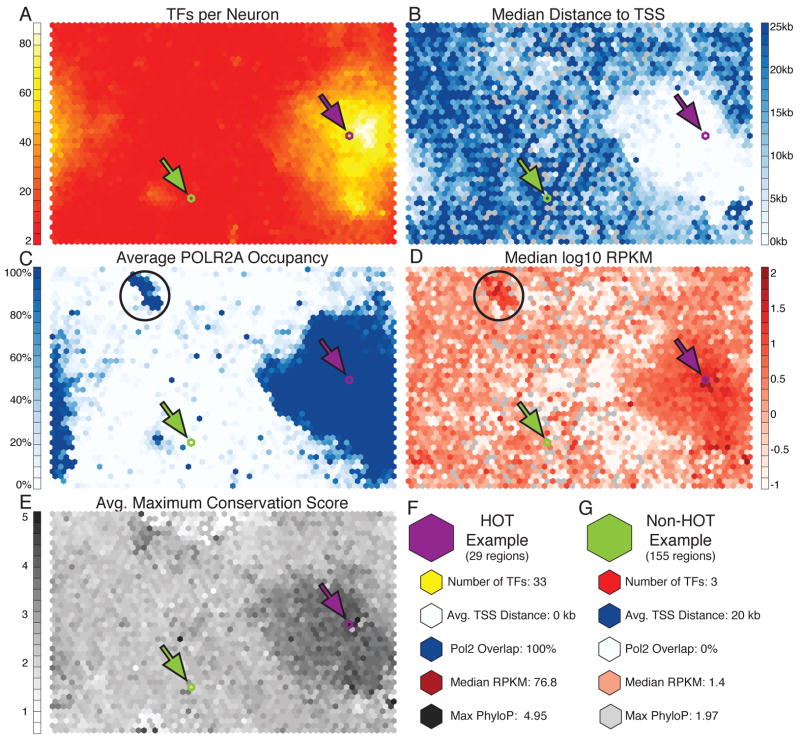
We identified co-localization patterns with an unusually high number of TFs (Fig. 3A). We consider the CLPs with more than half of the measured factors bound to be analogous to previously described HOT regions (Moorman et al., 2006). The CRMs in these regions have been found to be frequently associated with promoters and represent motif-less binding of TFs potentially to an open region of chromatin (Gerstein et al., 2010; Moorman et al., 2006; Roy et al., 2010; Yip et al., 2012). Accordingly, as shown in Fig. 3C, we show that the HOT regions largely overlap RNA Polymerase II binding and contain CRMs which are closer to the TSS than CLPs composed of fewer factors (Fig. 3B). Interestingly, using our SOM analysis, we also identify many TFs that co-localize with few partners and, thus, tend to avoid binding to HOT regions, which had not been reported previously. For example, the combination of CTCF+RAD21+SMC3 is known to act as an insulator and patterns that contain these factors are mostly excluded from the HOT regions. Other factors not binding to HOT regions include BACH1, CTCFL, MAFF, MAFK, NFE2, SETDB1, SPI1, USF1, USF2, and ZNF143.
Fig. 3.
General properties of co-localization patterns and HOT regions. (A) Overlay of the number of TFs comprising each CLP demonstrates that some CLPs represent binding of ‘HOT’ regions where a large number of factors bind the genome in close proximity. These regions are close to the TSS (B), have high POLR2A occupancy (C), drive high expression (D), and have higher maximum conservation than non-HOT regions (E). (F) An example of a CLP describing HOT binding (depicted by purple arrow in SOM plots). This CLP has 29 CRMs which match, consists of 33 different TFs binding, overlaps the TSS, has high POLR2A occupancy, has a very high RPKM, and as generally conserved. (G) A non-HOT region with only 3 TFs binding (depicted by green arrow in SOM plots). There are 155 CRMs matching this pattern with an average of >20kb from the TSS, no POLR2A, low RPKM, and lower maximum conservation. Black circles represent an interesting cluster of CLPs associated with high expression and only POLR2A with no other TF binding. See also Figure S3 and S4.
Analysis of RNA expression for targets of each of the CLPs reveals that HOT regions up-regulate gene expression, which is consistent with the observation that they overlap POLR2A (RNA Polymerase II) binding and the hypothesis that they locate at active promoters (Fig. 3D). Interestingly, we find a small cluster of highly expressed CLPs that are distinct from the HOT regions but show similar properties and are bound by only the POLR2A complex. Our results suggest that either these regions require very few TFs or that they belong to CLPs containing TFs not included in our data set (Fig. 3C–D, black circles). The sets of genes regulated by these CLPs are significantly enriched for mRNA processing, specifically ribosomal genes, mRNA splicing and transcription termination.
Another interesting and novel observation is that HOT regions have very different conservation patterns than those regions matching non-HOT CLPs. This is particularly evident in the comparison of minimum and maximum conservation scores where we observe that both the lowest and highest conservation scores correspond to the HOT CLP regions (Fig. 3E, Fig. S3). The observation suggests that HOT CRMs contain both very conserved and fast evolving components, which is in concordance with the theory that the binding site motifs are more conserved than background sequence but the arrangement of the motifs (copy number, order, orientation, spacing) evolve more rapidly (Xie et al., 2008).
We detail two CLPs in Fig. 3F–G to demonstrate the above differences between the two types of regions. Fig. 3F shows a CLP with 29 matching CRMs. These CRMs have 33 bound TFs, are on average at the TSS, all have POLR2A binding, have a very high median RPKM gene expression value of 76.8, and are conserved with a maximum PhyloP score of 4.95. This HOT region is contrasted with a non-HOT region in Fig. 3G where we demonstrate a CLP with 155 matching CRMs. These CRMs are approximately 20kb from a TSS, have no POLR2A overlap, have low expression of associated genes (1.5 median RPKM), and have lower maximum conservation with a PhyloP score of 1.97. This non-HOT region represents the canonical MAFF+MAFK+NFE2 binding complex (Igarashi et al., 1995).
DNase I Sites Overlap Only 60% of CRMs
We next compared the co-localization patterns with DNase I hypersensitive sites from ENCODE in K562 cells. DNase I hypersensitive sites were recently suggested to identify 95% of TF binding when pooled across a large number of cell types (The ENCODE Project Consortium, 2012). However, restricting our analysis to K562 cells, we found that many CLPs do not overlap DNase I sites and only 60% of CRMs have any overlap (Fig. S4). To rule out the possibility that the low overlap rate is a threshold artifact, we extended the published DNase I site peaks and found that about 35% of CRMs are located more than 1kb away and 22% of CRMs are located more than 5kb from the nearest DNase I peaks (Fig. S4). This discrepancy with previously reported numbers appears to be due to the high complexity of TF binding in promoter regions which are identified by the DNase regions and less complex patterns of binding in distal regions which are often independent of such sites. The overlap is significantly lower at distal binding regions for almost all TFs (Fig. S4). HOT regions, typically at promoters, are almost ubiquitously identified by the DNase I assays. Furthermore, the two different DNase I assays used by the ENCODE consortium (those from the University of Washington and from Duke University) appear to identify different subsets of CLPs with the Duke-developed assay more completely overlapping specific CLPs and the UW-developed assay more broadly identifying more CLPs (Fig. S4). Thus, our study reveals that many binding regions are only found by analysis of TF binding patterns and would be missed by DNase I hypersensitive sites assays.
We compared the CRMs with previously published chromatin state data from K562 cells (The ENCODE Project Consortium, 2012). We found that the CRMs that overlap with DNase I sites are enriched at active promoters, weak promoters, and strong enhancers. On the other hand, the CRMs that do not overlap with DNase I sites are significantly enriched for weakly transcribed regions, polycomb repressed regions, and heterochromatic regions. These findings suggest that TF binding and co-localization at more active regulatory regions that are readily identified by DNase I experiments but those in more silent regions are not. We further investigated the co-localization patterns that do not overlap with DNase I sites and found that previously reported heterochromatin-bound factors are in this category, such as SETDB1 and ZNF274 (Frietze et al., 2010; Schultz et al., 2002; The ENCODE Project Consortium, 2012). In fact, the CLP of SETDB1+TRIM28+ZNF274 has no DNase I overlap as shown by either assay but maintains a significant amount of GO and Pathway enrichment. However, only 5% of the non-DNase I overlapping CRMs contain heterochromatin-bound factors SETDB1 or ZNF274, suggesting large variety of TF co-localization outside of DNase I sites. The location of a CRM in open chromatin or heterochromatin may specify the distinct roles different CLPs play in the genome.
SOM Reveals Different Binding Patterns Across Conditions
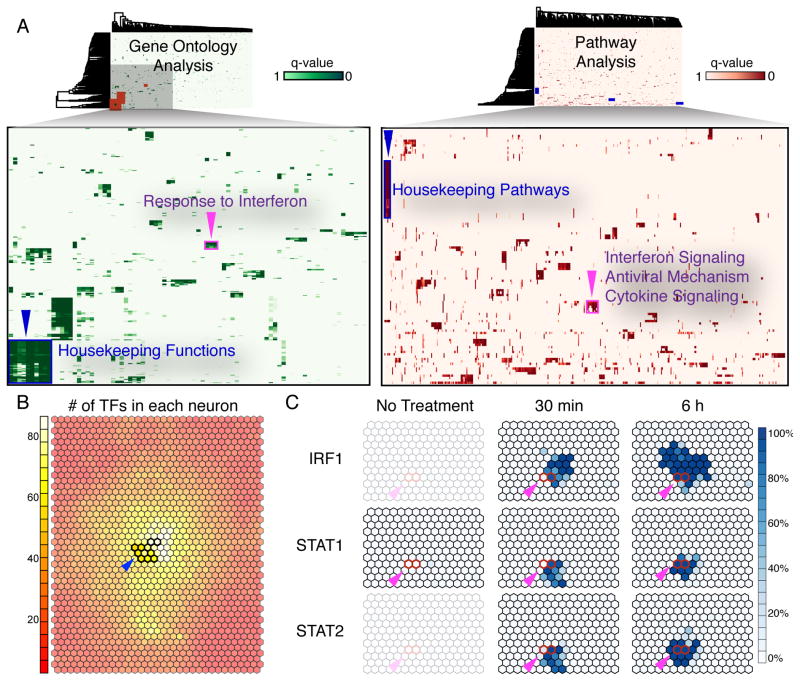
To further understand the functional implications of the co-localization patterns and their topological relationship on the SOM, we conducted systematic GO and pathway enrichment analysis on CRMs matching each CLP. We clustered the enriched functional terms based on the adjusted p-values of the enrichment (Fig. 4A). Strikingly, the largest clusters on both GO and pathway heatmaps are formed by CLPs located at the center of the HOT regions on the SOM (Fig. 4B). These CLPs are enriched with housekeeping GO terms and pathways suggesting the promoters of housekeeping genes are largely accessible and are bound by many different TFs.
Fig. 4.
GO and Reactome pathway analysis of all co-localization patterns. (A) The enriched GO and pathway terms for each CLP were clustered and show some common terms that exist for multiple CLPs. (B) HOT regions (blue arrows) are significantly enriched for GO and pathway terms related to housekeeping functions. (C) A cluster enriched for immune function (purple arrows) is also enriched for STAT1, STAT2, and IRF1 binding. This cluster includes regions which are activated under interferon response in our data. The red outlined CLPs indicate a set of CRMs bound by IRF1, STAT1, and STAT2 after 30 minutes of interferon treatment (left) and after 6 hours of interferon treatment (right). STAT1 binding can be shown to only exist after interferon treatment.
We also examined the dynamics of CLPs under different experimental conditions which had not been systematically investigated previously. We analyzed a series of ChIP-seq data for STAT1 and IRF1 after treatment with interferon (Fig. 4C). We found time dependent association among IRF1, STAT1, and STAT2. Specifically, we identify one binding pattern representing IRF1+STAT1+STAT2 binding 30 minutes after interferon treatment, and another representing IRF1+STAT1+STAT2 binding at both a 30 minute and a 6 hour time point. Accordingly, the enriched GO terms for CRMs matching these two CLPs include many interferon response and immune response related terms, and the enriched pathways also include interferon signaling and innate immune system. Our results demonstrate that complex patterns of TF co-localization can change dynamically over short temporal periods and in a functionally relevant fashion.
Regulatory Changes Across Cell Types
Because the inputs to the SOM are regions with TF binding, we are able to include binding patterns from multiple cell types which have ChIP-seq data from the same TFs. This analysis allows us to explore co-localization patterns that may be unique or shared among cell types. We trained two multi-cell-type SOMs to examine the variance of TF association relationships between different cell types. The first multi-cell-type SOM is trained with CRMs from two cell types: K562 and GM12878. ChIP-seq data for 53 TFs assayed on both cell lines were included in this comparison and we term it “deep comparison”. The second multi-cell-type SOM is trained with CRMs from five cell types: K562, GM12878, H1, HepG2, and HelaS3. ChIP-seq data for 19 TFs were shared in all the five cell lines and included in the comparison; we term this analysis the “broad comparison”.
In the “deep comparison”, our analysis revealed that approximately one third of the co-localization patterns are K562-specific, one third of the patterns are GM12878 specific, and one third of the patterns are shared by two cell types (Fig. 5). An intriguing feature of the “deep comparison” is that the patterns specific to one cell type are mostly clustered together, forming the “territory” of that cell type. This phenomenon is likely to reflect the different preferences of TF usage and TF co-localization between cell types. Patterns that are shared by the two cell types represent TF-associations that perform similar function in the two cell types. For example, REST functions as a neuronal gene repressor in both K562 and GM12878 and its co-localization with ZNF143 is shared in the two cell types. The REST+ZNF143 CLP is also enriched with neuronal activity functional terms for both cell types. Other shared patterns include SPI1+ELF1, SIN3A+MAX+MXI1, EP300+PML+SPI1 and MAZ+EGR1+SP1, indicating that these may be common co-localizations.
Fig. 5.
A K562 and GM12878 SOM can be generated using factors shared between the two cell types. The CLPs are shaded based on the number of CRMs from either cell type matching each CLP with GM12878 CRMs shaded red and K562 CRMs shaded blue. The CLPs containing a mixture of the two cell types have an intermediate shading as depicted in the legend. A large number of the CLPs are cell-type specific and the HOT regions (region outlined in yellow) overlap the cell-type specific CLPs. These HOT CLPs are enriched for housekeeping GO terms. We also display cell-type specific GO terms that are not in HOT regions as well as examples of common CLPs such as REST+ZNF143, ZNF143+CTCF+SMC3+RAD21, and CTCF+RAD21. See also Figure S5.
Cell type specific patterns are likely to account for the phenotypic difference between cell lines. A majority of the HOT patterns are cell type specific, which supports observations from previous studies (Yip et al., 2012). As above, these HOT regions are enriched for housekeeping functional GO terms and pathways. Although these enrichments appear in cell-type specific CLPs, the terms are not necessarily specific to one cell type. This is because different CLPs in the two cell types can describe the same genomic binding region and, thus, two different CLPs can be regulating the same sets of genes. The cell type-specific CLPs are also enriched with functional terms that are associated with the properties of the individual cell types. For example, lymphocyte co-stimulation and regulation of antigen processing and presentation are enriched in CLPs specific for GM12878, a lymphoblastoid cell line. Furthermore, tumor necrosis factor receptor binding and apoptotic execution phase are enriched in K562-specific CLPs, which have been previously reported as enriched in K562-specific gene expression (Hietakangas et al., 2003).
The “broad comparison” across five cell types also results in patterns specific to each cell type (Fig. S5). This is surprising given the limited number of TFs that are shared across the cell types. However, the separation of co-localization patterns into cell-type specific domains underscores the different TF utilization patterns apparent in the “deep comparison”. Furthermore, these domains are enriched for GO and pathway terms that again match housekeeping terms in HOT regions and functional terms specific to each cell-type in the cell-type specific domains. We also identify HOT binding patterns that are shared among cell-types, likely due to the limited scope of factors used.
Co-Binding Mediated by TF Protein-Protein Interactions
The co-localization patterns that we identify may be due to stable physical interaction of the proteins (directly or indirectly) as part of the same complex whereas others may be due to co-localizations that occur only in the context of the regulatory DNA. In order to further explore the potential mechanism of co-localization, a mapping of protein-protein interactions was integrated with the CLPs. Many efforts have been carried out to construct a comprehensive picture of the protein interaction networks to understand the regulation of biological processes in the cell (Malovannaya et al., 2011; Rual et al., 2005; Stark et al., 2006; Stelzl et al., 2005). However, these datasets are derived from literature curation, generated by using yeast two-hybrid system, or performed in a different cell type. Because TF co-bindings are highly context dependent and cell-type specific, these previously defined networks provide an inaccurate and incomplete picture in K562 cells. We identified endogenous protein complexes from K562 using antibody immunoprecipitation and mass spectrometry (IP-MS) to identify potential TF protein-protein interactions that likely occur in vivo (Table S1). A total of 24 antibodies used for ChIP-seq assays were tested which enabled us to investigate TF-TF interactions in our dataset that occur in the same protein complexes. We identified 40 pairs of TF-TF interactions for which we have ChIP-seq data for both TFs. Among the 40 pairs, 7 (17.5%) were previously known protein-protein interactions, whereas the remainder are novel. Importantly, we found that 30 out of the 40 TF-TF interactions overlap with the co-localization patterns that we identified with ChIP-Seq experiments (p-value <0.05).
To further examine the scenario where two TFs are tethered by a third protein, we also included 26 TF IP-MS datasets that were performed in K562 cells but not used for the ChIP-seq assays. With the interactions identified by the total of 50 antibodies, we constructed a protein-protein interaction (PPI) network that revealed both direct and indirect interactions between TFs studied in our ChIP-seq dataset. In total, we identified 207 direct or indirect interactions and 172 (83%) of them match a co-localization pattern in our SOM (p-value <0.05). The TF co-binding patterns suggested to be due to protein-protein interactions are members of ~40% of the CLPs. In addition, we were able to associate some CLPs directly with PPI sub-networks where several TFs contained in the co-localization pattern are connected (Fig. 6). These associations reveal co-localization patterns that are due to a protein complex rather than simply individual binding events on the DNA.
Fig. 6.
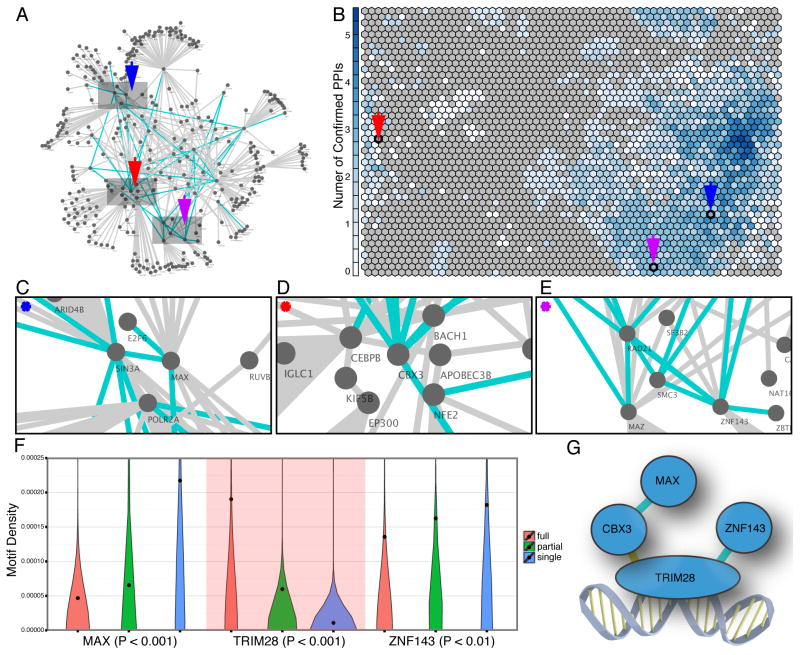
Association between SOM trained from ChIP-seq data and PPI constructed from mass spectrometry data. (A) A PPI network showing both direct and indirect TF interactions was constructed based on the mass spectrometry assays of immunoprecipitation of 50 TFs. The blue edges in the network represent direct TF interactions identified by the IP-MS data and the grey edges represent indirect TF interaction. (B) Many TF interactions are identified in the co-localization patterns discovered by the SOM. The numbers of TF interaction pairs seen in the SOM are represented by the blueness of each CLP. Co-localization patterns represented in each CLP are mapped to a subnetwork on the PPI network. For example, (C) The CLP pointed by the blue arrow is mapped to the subnetwork consisting of E2F6, SIN3A, MAX, and POLR2A; (D) The CLP pointed by the red arrow is mapped to the subnetwork consisting of CBX3, BACH1, NEF2, CEBPB, and EP300. KIF5B serves as an intermediate protein tethering CEBPB and EP300; (E) The CLP pointed by the purple arrow is mapped to the subnetwork consisting of RAD21, MAZ, SMC3, and ZNF143. SF3B2 serves as a mediator tethering MAZ and ZNF143; (F) Violin plot of the motif usage for MAX, TRIM28, and ZNF143 when they all co-localize (red), a subset co-localize (green), or bind independently (blue). The dots are average motif density in each category. MAX and ZNF143 have higher motif density when they bind independent of the complex. TRIM28 (background shaded red) has higher motif density when it binds as a protein complex and is likely to interact with the DNA in this complex; (G) An illustration of the MAX, TRIM28, ZNF143, CBX3 protein complex identified by the IP-MS dataset showing the predicted interaction with the DNA. Direct interactions are in blue while the yellow interaction represents an indirect interaction in our data that has been previously validated. See also Table S1.
The binding of a protein complex to the DNA sequence may be primarily through a subset of the factors acting as a DNA binding anchor to which the other protein components are tethered. We explore this possibility by examining motif usage which is suggestive of direct TF interaction with DNA. For members of complexes suggested by the IP data, we compared the motif usage for TFs within a CLP that contains multiple interacting factors and with CLPs where the factor is not found with these partners. We found uneven motif usage is evident in many of these cases. For example, the CLP that consists of MAX, ZNF143, TRIM28, and CBX3 was present as a connected PPI sub-network in the IP-MS data: we identified direct interactions between MAX-CBX3 and TRIM28-ZNF143 as well as an indirect interaction between CBX3-TRIM28 that was also identified in previous studies (Higo et al., 2010; Rosnoblet et al., 2011; Ryan et al., 1999). For the three TFs for which we have known DNA binding motifs (MAX, ZNF143, and TRIM28) there was a significant difference of motif usage (Fig. 6F). MAX and ZNF143 have significantly higher motif density when they bind in CLPs independent of the other partners in this complex. This trend is evident in all versions of motif consensuses for the two TFs. In contrast, TRIM28 showed higher motif density when it binds together with the other TFs in the complex than when independent of the complex. The results suggested that when the four TFs form a complex their binding to the DNA sequence is likely anchored through TRIM28, while MAX and ZNF143 are likely to be tethered (Fig. 6G). Thus, the combination of SOM and PPI information can be used to decipher binding relationships among the different members of protein complexes.
Discussion
How the thousands of TFs in human cells co-localize under different conditions is a central question in understanding gene regulatory mechanisms. To better understand this TF co-localization, two complimentary efforts are needed. First, we need a comprehensive map of TF binding sites in different cell types and conditions. In this work, we aimed to interpret the most comprehensive human ChIP-seq dataset to date that is comprised not only of the largest number of TFs but also the richest of cell conditions. Second, we need powerful computational methods to thoroughly and elegantly interpret the large datasets. The exponentially increasing number of high-throughput datasets has provided an unprecedented opportunity to study the complexity of TF co-localization relationships with their many thousands of targets in the genome, but the large volume and high dimensionality has made the data unintuitive to understand and difficult to interpret without advanced computational methods. The application of SOMs in this method provides an elegant way to not only explore these complex relationships in a comprehensive and rapid fashion but also to visually interpret the results. This work represents a significant advance over previous studies that study the co-localization of two TFs under the context of a third TF, a more limited solution (Gerstein et al., 2012). The SOM method allows all combinatorial associations to be explored.
The advantage of analyzing TF co-localization in higher dimension is that it allows much more insight to the complexity of binding that cannot be captured by previous methods. For example, in the FOS-NFYB co-localization analysis, we revealed more scenarios of co-localization between FOS and other TFs than previously known. We also showed that a majority of TF co-localization involves canonical binding contexts coupled with additional complex binding patterns, which often associate with different regulatory outcomes. These data further prove that the previous view of TF co-localization in a low dimension is an over-simplification. Another advantage of our method is that it is highly integrative, which naturally enables cross-condition analysis and allows convenient investigation of many properties of TF co-localization (e.g. associations with gene expression levels, distance from promoters, etc.) by integrating other functional genomics data that are abundantly available.
The integration of two major high-throughput fields, genomics and proteomics, is another advance in this study. We incorporate a large ChIP-seq dataset and a large endogenous protein-protein interaction dataset from the sample cell line to both cross-validate one another as well as explore the mechanism of TF co-localization. In general, there are two canonical models to explain TF co-localization. First, TFs co-localize because their binding motifs locate near each other on the genome sequence with little or weak association in as part of a protein complex. Alternatively, TFs co-localize because they interact in a stable protein complex. From the study of TF binding motifs, it is likely that the first model only partially explains TF co-localization as many of the TF binding sites do not contain binding motifs (e.g. Martone et al., 2003). Although far from complete, our IP-MS dataset coupled with the SOM data from the large ChIP-seq dataset allowed us to take these observations to a new level. We found that for a TF complex deduced from IP data, only some components within a CLP have a motif and likely contact the DNA; other components are not enriched even though the motifs may be enriched in other CLPs. Thus, by using the SOM map and IP complex data, we can propose binding relationships among the different factors at many regions of the genome, thereby providing a better understanding of likely mechanisms of binding. We expect that with larger and more comprehensive data available in the near future we will be able to define an even better map of the relationships that govern the complex mechanisms controlling gene regulation.
Experimental Procedures
ChIP-seq Data
All analysis is performed on the GRCh37 (hg19) reference genome. ChIP-seq experiments protocol, quality control, and preprocessing followed ENCODE standards and were performed as part of the ENCODE standard processing pipeline(Landt et al., 2012; The ENCODE Project Consortium, 2012). All data are available for download from the UCSC ENCODE portal. Peak regions identified from different ChIP-seq data for the same TF in identical cell types and conditions were merged into a union set for our downstream analysis.
Determination of Cis-Regulatory Modules (CRMs)
We collected and standardized 158 ChIP-seq data sets representing the binding of 128 TFs in different conditions of K562 cells. We also collected 52 pairs of ChIP-seq data sets in K562 and GM12878 cells for the “deep comparison” and 18 sets of ChIP-seq data in K562, GM12878, HepG2, HelaS3, and H1hESC cells for the “broad comparison”. Using these data, we defined a cis-regulatory module as the maximum overlapping block of the intersection of all TFs binding peaks and required that at least 2 TFs bound in a CRM to be considered for further analysis. In the “deep comparison” and “broad comparison” CRMs were defined as the maximum overlapping block of TFs in an individual cell-type.
In addition to the intersection method used, we explored the use of 500bp windows, 1kb windows, and DNase I hypersensitive sites to define cis-regulatory modules.
We found that using window approaches resulted in significantly more CRMs than the other two approaches because most CRMs were likely broken into multiples due to the windowing. Furthermore, we found that using the intersect approach resulted in approximately the same number of CRMs as using DNase hypersensitive sites (approximately 150,000 in the “deep comparison” and 280,000 in the “broad comparison”) while allowing us to identify CRMs which may be independent of DNase I sensitivity.
SOM training and parameters
We identified each cis-regulatory module as either bound (1) or not bound (0) by overlap with peaks from each TF. This results in the cis-regulatory modules being represented as a binary vector of 128 dimensions with each dimension representing a TF. These vectors are used as input to the SOM and resulting descriptions of each neuron are also described in this form.
For each SOM trained, we followed the following rules:
The SOM is initialized as a random toroid.
The SOM is hexagonal.
- The total number of neurons is:
- The number of neurons along the y-axis is:
- The number of neurons along the x-axis is:
The SOM is trained for 100 epochs (that is, complete iterations through the data set)
The SOM update radius was ⅓ of the map size with a learning rate (alpha) of 0.05. These were linearly decreased throughout the training process.
We selected for analysis the best of 1000 trials based on lowest quantization error (defined as the average Euclidean distance of all CRMs to their best matching neuron).
The number of neurons is a modification of the simple heuristic (5 × sqrt(k)) proposed by (Vesanto, 2005) where we better account for the total number of dimensions of our larger data set as well as the number of training samples. Our modification consists of the following two adjustments. 1) Parameter k scaled to account for “effective” size of input CRMs by scaling to the size of the smallest input (only for multiple cell-type comparisons). 2) Replacing the constant ‘5’ with a parameter to scale for the dimensionality being explored by the SOM. The rationale here being that a SOM with higher dimensionality will likely expand over a larger space and will require more neurons to properly model the distribution.
The training described above is performed in R using a variant of the ‘kohonen’ package available from CRAN. Minor modifications were performed to the R package to allow for better handling of the large data sets in memory. Furthermore, significant changes to the graphical output of the package were made to allow for the improved figures displayed here and on the supplemental web site. Final optimal seeds for the training were K562 SOM: 810, “deep comparison” SOM: 293, and “broad comparison” SOM: 272. 100 epochs of training resulted in stabilization of the SOMs and of the 1000 iterations of the SOM there was minimal divergence with the best SOM having less than 0.3% difference in error than the average error of the non-optimal SOMs. Final SOM sizes were 62×46, 50×40, and 47×34 for K562, “deep comparison”, and “broad comparison” respectively and average CRM distance to the best matching neuron was 2.45, 1.38, and 0.22 for K562, “deep comparison”, and “broad comparison” respectively.
The input for multiple cell type SOM training consists of CRMs identified from multiple genomes. For example, in the “deep comparison” SOM, the CRMs from K562 cells and GM12878 cells were identified independently and pooled together for the training. After the training, the CRMs from the two cell types with the sample CLP were clustered together. Since we know the origin of each CRM, we could calculate the proportion of the same CRMs between the two cell types.
TF pull down assays
For each immunopricipitation (IP) experiment, 2 X 108 of frozen K562 cells were thawed in 12 ml cold PBS at 4°C for 1 hour on neutator. The cells were spun at 1,500 rpm for 3 minutes, and the supernatant was removed. Then pellets were suspended in hypotonic buffer, and dounced by homogenizer on ice for 30 strokes. The lysates were aliquoted into two tubes, and centrifuged at 600 g at 4°C for 8 minutes. The supernatant was discarded, nuclear pellets were resuspended in 1ml 1X RIPA buffer and incubated on ice for 30 min. The nuclear lysates were further centrifuged at 14,000 rpm at 4°C for 15 min. The supernatant was transferred to a 50 ml falcon tube, and the total volume was adjusted by 1X RIPA buffer to a final of 30 ml. Each tube was supplied with 12 ug antibody (or equal amount of normal IgG in a parallel control sample), and incubated at 4°C with neutator rocking overnight.
Each sample lysates were mixed with 150 μl of prewashed Protein A/G-agarose beads, and then incubated at 4°C for 1 hour on neutator rocker. Agarose beads were pelleted and washed with ice-cold RIPA buffer three times and ice-cold PBS once. The beads were transferred to a 1.5 ml eppendorf tube, and resuspended in 55 μl of 2X Laemmli buffer containing beta-mercaptoethanol, boiled and stored in −20 °C for further usage.
Constructing PPI networks
Among the 50 TF antibodies used for IP-MS experiments, 24 TFs also have corresponding ChIP-seq data in K562 cells. Besides the targeted TF, the IP sample also pulled down other proteins with average spectral counts greater than that in the parallel control sample. Among this data set, there are 40 pairs of TF protein-protein associations in which both proteins have corresponding ChIP-seq datasets in K562 (defined as direct PPI). To preserve weak protein interactions during complex isolation, we constructed a protein-protein network to discover more potential protein-protein associations. After pooling all the 50 TF IP-MS data sets together, we scored protein-protein interactions based on spectral counts using Significance Analysis of INTeractome (SAINT) software package (Choi et al., 2012). Protein-protein associations with average probability greater than 0.7 and containing at least one TF which has ChIP-seq data in this study were selected and combined with the previous direct TF PPI, yielding a complicated TF protein-protein association network (Table S1). Protein-protein associations tethered by a third protein in the network were defined as indirect PPI. For example, if protein B and C are identified from mass spectrometry analysis of protein A IP, B and C are considered as indirect PPI.
For each of the direct and indirect TF interactions identified by mass spectrometry data, if both TFs are contained in a significant trans-binding pattern in SOM, we consider the interaction being cross validated. A trans-binding pattern consisting of more than two TFs has multiple possible protein-protein interaction pairs, and therefore can validate more than one TF interactions (Figure 6B). 172 out of 207 interactions (both direct and indirect) are cross-validated using the above method. We calculated the p-values by fixing the pulled down TFs while permuting all the partners identified by mass spectrometry and calculating the odds of getting higher cross validations. A total of 200 permutations were performed, enabling us to estimate the p-value to the level of 0.05.
Supplementary Material
Highlights.
Co-localization patterns of 128 TFs in human cells
A novel application of SOMs to study high-dimensional TF co-localization patterns
Co-localization patterns are dynamic through stimulation and across cell types
Many TF co-localizations can be explained by protein-protein interaction
Acknowledgments
We thank the ENCODE consortium for providing data used in these analyses. This project is funded by NHGRI grand U54HG004558.
D.X., A.P.B., and M.S. conceived the research plan. L.W. and M.S. designed mass spectrometry experiments. D.X. and A.P.B. performed computational analysis. L.W. J.Z. and T.K. performed experiments. L.W. performed mass spectrometry data analysis. D.X., A.P.B. and M.S. wrote the manuscript.
References
- Balázsi G, Barabási AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina MD, Troyanskaya OG. An effective statistical evaluation of ChIPseq dataset similarity. Bioinforma Oxf Engl. 2012;28:607–613. doi: 10.1093/bioinformatics/bts009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Liu G, Mellacheruvu D, Tyers M, Gingras A-C, Nesvizhskii AI. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr Protoc Bioinforma Ed Board Andreas Baxevanis Al. 2012;Chapter 8(Unit 8.15) doi: 10.1002/0471250953.bi0815s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013 doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3’ ends of ZNF genes. Plos One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Poukkula M, Heiskanen KM, Karvinen JT, Sistonen L, Eriksson JE. Erythroid differentiation sensitizes K562 leukemia cells to TRAIL-induced apoptosis by downregulation of c-FLIP. Mol Cell Biol. 2003;23:1278–1291. doi: 10.1128/MCB.23.4.1278-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo S, Asano Y, Kato H, Yamazaki S, Nakano A, Tsukamoto O, Seguchi O, Asai M, Asakura M, Asanuma H, et al. Isoform-specific intermolecular disulfide bond formation of heterochromatin protein 1 (HP1) J Biol Chem. 2010;285:31337–31347. doi: 10.1074/jbc.M110.155788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Conditional expression of the ubiquitous transcription factor MafK induces erythroleukemia cell differentiation. Proc Natl Acad Sci U S A. 1995;92:7445–7449. doi: 10.1073/pnas.92.16.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T. Self-Organizing Maps. Berlin, Heidelberg, New York: Springer; 2001. [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA. Homotypic regulatory clusters in Drosophila. Genome Res. 2003;13:579–588. doi: 10.1101/gr.668403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. Distribution of NF-KB-binding sites across human chromosome 22. Proc Natl Acad Sci U S A. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnoblet C, Vandamme J, Völkel P, Angrand PO. Analysis of the human HP1 interactome reveals novel binding partners. Biochemical and Biophysical Research Communications. 2011;413:206–211. doi: 10.1016/j.bbrc.2011.08.059. [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesanto J. SOM Toolbox: implementation of the algorithm 2005 [Google Scholar]
- Xie D, Cai J, Chia NY, Ng HH, Zhong S. Cross-species de novo identification of cis-regulatory modules with GibbsModule: application to gene regulation in embryonic stem cells. Genome Res. 2008;18:1325–1335. doi: 10.1101/gr.072769.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.