Abstract
Despite recent progress on estimating the heritability explained by genotyped SNPs (hg2), a large gap between hg2 and estimates of total narrow-sense heritability (h2) remains. Explanations for this gap include rare variants, or upward bias in family-based estimates of h2 due to shared environment or epistasis. We estimate h2 from unrelated individuals in admixed populations by first estimating the heritability explained by local ancestry (hγ2). We show that hγ2 = 2FSTCθ(1−θ)h2, where FSTC measures frequency differences between populations at causal loci and θ is the genome-wide ancestry proportion. Our approach is not susceptible to biases caused by epistasis or shared environment. We examined 21,497 African Americans from three cohorts, analyzing 13 phenotypes. For height and BMI, we obtained h2 estimates of 0.55 ± 0.09 and 0.23 ± 0.06, respectively, which are larger than estimates of hg2 in these and other data, but smaller than family-based estimates of h2.
Introduction
Understanding the genetic architecture of complex human phenotypes is a fundamental question to the field of genetics, with broad implications for identifying genes related to disease and predicting individual risk profiles1-6. A central element of this problem is estimating narrow-sense heritability (h2), the fraction of phenotypic variation in a population determined by genetic variation under an additive model7. While the last decade of genome-wide association studies (GWAS) produced thousands of novel loci associated with hundreds of phenotypes8, the sum of their effects ( ) explain only a small fraction of the estimated heritability for most phenotypes5. The gap between and h2 is called the “missing heritability” and several explanations for this difference have been posited, including upward bias in estimates of h22,4,9. The objective of this work is to develop a method for estimating h2 (defined in Methods) that (1) does not require closely related individuals, (2) can be applied to both quantitative and case-control phenotypes, and (3) is able to localize narrow-sense heritability to individual chromosomes or other genomic segments.
Current approaches to heritability estimation proceed by phenotyping many closely related individuals with a known genetic relationship, such as monozygotic (MZ) and dizygotic (DZ) twins7. Yang et al.10 avoided the use of related individuals by applying linear mixed models to estimate the heritability explained by genotyped SNPs (hg2). hg2 corresponds to the fraction of phenotypic variation that could be captured by under an additive model if GWAS sample sizes were infinitely large. While current estimates of hg2 are often much larger than , they are typically only slightly more than half of h2 11. One reason hg2 is less than h2 is because hg2 does not include the contribution of variants poorly tagged by the genotyping platform, such as rare variants. Another reason for the difference in heritability estimates is that existing methods for estimating h2 can be biased12,13, since they rely on related individuals. As a result, epistatic interactions between SNPs, gene environment interactions, and the shared environmental factors of related individuals can all lead to inflated estimates of h2 12,13. We recently showed that by jointly using related and unrelated individuals it is possible to obtain less biased estimates of h2 11. However, the joint fit will still lead to inflated estimates of h2 in the presence of shared environment11, and can not be applied to case-control phenotypes.
In this work we propose a new approach for estimating h2, which takes as input the phenotypes and genotypes of admixed individuals such as African Americans. We show via analytical derivation as well as extensive simulation over both simulated and real genotype data that heritability explained by local ancestry (hγ2) is related to the total narrow sense heritability h2 via the equation hγ2 = 2FSTCθ(1−θ)h2, where FSTC is a specific measure of weighted allele frequency differences between ancestral populations at causal loci (see Online Methods) and θ is the fraction of European ancestry14,15. Since our approach does not use closely related individuals it is free from bias due to epistasis, gene environment interactions, and shared environment effects. Unlike previous work in which h2 estimates could not be obtained for case-control phenotypes11, our current approach can obtain estimates of h2 for both quantitative and case-control phenotypes, achieving goals (1) and (2). Furthermore, unlike previous methods that provide genome-wide estimates, we are able to estimate h2 for a particular genomic region, such as a chromosome, achieving goal (3). Our approach can be applied to all existing and future GWAS of admixed populations, without requiring additional expensive and time-consuming collections of large numbers of MZ and DZ twins.
We applied this approach to 21,497 African Americans from the NHLBI CARe, WHI-SHARe, and AAPC projects, analyzing 12 quantitative phenotypes and 1 case-control phenotype. For height and BMI, we obtained h2 estimates of 0.55 ± 0.09 and 0.23 ± 0.06, respectively, which are larger than estimates of hg2 in these and other data sets but smaller than twin-based estimates of h2, consistent with inflation in twin-based estimates because of shared environment or epistasis. We also estimated the heritability of height for each chromosome and found a significant correlation between chromosome length and heritability (p-value < 0.003).
Results
Overview of method
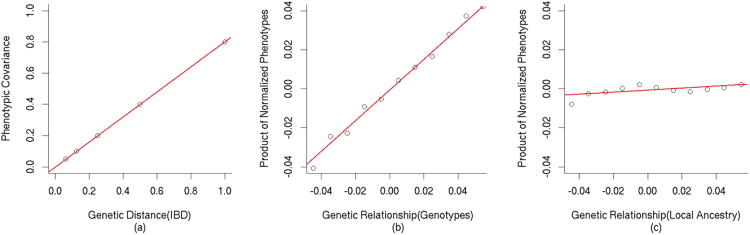
We consider three approaches to estimating heritability for a phenotype with a narrow-sense heritability of 80%. First, the classic approach to estimating heritability is to divide the phenotypic covariance of related individuals by the fraction of the genome they share IBD13. In this instance, the phenotypic covariance of pairs of related individuals will be 0.80 times the fraction of genome shared IBD (Figure 1a). The second approach, developed by Yang et al.10, is to estimate the genetic relationship of unrelated individuals over genotyped SNPs and applied a linear mixed model with the genetic relationship matrix to estimate phenotype. To illustrate this approach we simulated 2 million independent pairs of individuals, regressing their normalized genetic similarity over the product of their normalized phenotypes giving a regression coefficient of 0.79±0.014 (Figure 1b). This Haseman-Elston regression16 shows how genetic similarity of unrelated individuals can be used to estimate heritability of genotyped SNPs (hg2). In general, the heritability explained by genotyped SNPs is less than the total narrow-sense heritability (h2) since phenotypic variation determined by poorly tagged SNPs such as rare variants will not be captured10.
Figure 1.
Relationships between genetic distance and phenotype for a trait with heritability = 80%. (a) The phenotypic covariance of pairs of individuals at different expected fractions of genome shared IBD is 0.8*%IBD. (b) Regression of genetic distance estimated from genetic variation against the product phenotypes normalized to have mean 0.0 and variance 1.0 has coefficient 0.79 (se = 0.014). (c) Regression of genetic distance estimated from local ancestry variation against normalized phenotypes has coefficient 0.033 (s.e. = 0.007) ≈2FSTCθ(1−θ)h2= 0.032, corresponding to h2 = 0.83 (s.e. = 0.18).
The approach used in this work is similar to that of Yang et al.10, but instead of using genotypes to estimate genetic similarity we use the number of copies of local ancestry in an admixed population. A crucial element of our approach is that the phenotypic variation described by variation in local ancestry ( ) is a function of all causal variation, not just that tagged by SNPs on the genotyping platform. This is because local ancestry tags both common and rare variation. To illustrate this approach we simulated 4 million unrelated admixed individuals from ancestral populations with genetic distance FSTC= 0.08 and an equal proportion of ancestry from each ancestral population θ = 0.5 (see Online Methods). Applying Haseman-Elston regression to regress the product of normalized phenotypes against genetic similarity of local ancestry, we observe a regression coefficient 0.033±0.007 ≈ 2FSTCθ(1−θ)h2= 0.032, corresponding to h2 = 0.83 (s.e. = 0.18) (Figure 1c). The Haseman-Elston regression used in generating these figures is for illustrative purposes (as in Figure 3 of [10]). In practice, we use a mixed model approach due to its lower standard errors10.
We first construct a local ancestry based kinship matrix Kγ, which is constructed similarly to the genotype-based kinship matrix K in previous methods10, but with local ancestry substituted for genotypes at each SNP. We use a variance components approach to estimate the phenotypic variance explained by variation in local ancestry ( ) and the residual phenotypic variance ( )10,17. We included genome-wide ancestry proportion θ and the top five principal components as fixed effects when fitting the mixed model (see Online Methods). The heritability explained by local ancestry is given by . Finally, to estimate h2, we use the formula hγ2 = 2FSTCθ(1−θ)h2, where FSTC is a specific measure of weighted allele frequency differences between ancestral populations at causal loci (see Online Methods). For dichotomous phenotypes we applied the same approach, but converted the observed scale estimates to a liability scale estimate of heritability using [18], and the published disease prevalence in African Americans. In our previous work11, this conversion was not possible because non-randomly ascertained individuals in multiple relatedness classes (e.g. siblings, first cousins, avuncular) were studied, and there is currently no method for accounting for ascertainment in such complex pedigrees. A complete description of the approach, along with an analytical derivation, is given in Online Methods.
Simulations with Simulated Genotypes
We first verified the analytical derivations and examined the properties of the approach under a simple simulation framework. We simulated the genotypes and local ancestry of 4,000 unrelated diploid individuals at 1,000 SNPs from a two-way admixed population with causal variant genetic distance FSTC, and either normally or uniformly distributed ancestry proportion θ. Each local ancestry segment contained exactly one SNP and all segments were generated independently. Phenotypes were simulated under an additive model with heritability h2 in which a proportion r of the 1,000 SNPs was causal (see Online Methods). We applied our method to estimate heritability over a range of values of FSTC, θ, r, and h2. For each parameter setting we estimated heritability from 2,000 independent simulated data sets. The results shown in Table 1 show that our heritability estimates are accurate across a range of parameter settings, confirming our analytical derivation. Results for additional parameter settings are shown in Supplementary Table 1.
Table 1.
Results of local ancestry based heritability estimation from simulated genotypes and simulated phenotypes over a range of population and disease architectures. Mean heritability estimates and standard errors are reported from 2,000 simulations for each choice of parameters.
| h2 | FST | r | ĥ2 |
|---|---|---|---|
| 0.8 | 0.30 | 1.0 | 0.802(0.003) |
| 0.8 | 0.30 | 0.1 | 0.802(0.005) |
| 0.8 | 0.15 | 1.0 | 0.800(0.005) |
| 0.8 | 0.15 | 0.1 | 0.804(0.006) |
The results also demonstrate the relationship between and the parameters FSTC, θ, and h2. For a fixed value of r, phenotypes with a larger h2 will have larger genetic effects resulting in larger . When ancestral populations are genetically distant (larger FSTC), variants are more likely to have a different frequency in the ancestral populations resulting in a concomitant increase in . Increasing the variance of θ results in a larger standard error around the heritability estimates.
Simulations with Real Genotypes
We made several simplifying assumptions in the above simulations that do not hold in real data sets. These include a single SNP per ancestry block, no genotyping error, no local ancestry inference error, no LD, a normal or uniform distribution of ancestry proportion, continuous phenotypes, and that the effect size distribution of common and rare variants used in computing FSTC was identical. To address these complexities, we took the approach of using real genotypes and simulating phenotypes. We simulated continuous and case-control phenotypes over 5,129 individuals (excluding close relatives) from the CARe cohort (see Online Methods). Although phenotypes were generated from SNPs sampled across all genotyped SNPs, we only used local ancestry information from every 5th SNP.
We tried a range of parameters for h2. Instead of simulating phenotypes under an infinitesimal model, we sampled a proportion of causal variants r. We could not alter ancestry proportion θ, since this is fixed in the real data set. However, we altered the effect size distribution of SNPs according to their value of FSTC.
The data did not contain a sufficient number of genotyped variants that were rare in both populations to simulate rare versus common effects. Instead we examined SNPs common in both populations (common) vs. SNPs rare in at least one population (uncommon). Only common variants were used in constructing the kinship matrix, and so uncommon variants will only contribute to hg2 via LD. The common SNPs had an FSTC of 0.15, while the uncommon SNPs had an FSTC of 0.25. We simulated phenotypes with a different proportion of phenotypic variance from uncommon variants (α). When α is different from 0, the kinship matrix variant and causal variant frequencies are different. The results in Table 2 show that simulations involving a large proportion of causal variants not included in the kinship matrix (high α) had a lower value of hg2 than h2 because the common variants did not completely capture the phenotypic variance driven by the uncommon variants. The parameter α also determines the study wide FSTC according to FSTC = (0.15(1-α) + 0.25α) (see Online Methods). The results shown in Table 2 use the correct value of α, and hence the estimates of h2 are unbiased. However, if we incorrectly assume that α=0 when it does not, then h2 will be biased by factor of (0.15(1- α) + 0.25α)/0.15. We describe this (and other potential sources of bias) in detail in the Discussion.
Table 2.
Results of heritability simulations over 5,129 African American individuals from the CARe cohort. Average estimates and standard errors of heritability explained from genotyped SNPs( ), and our local ancestry based estimate of heritability explained from all SNPs (ĥ2) are reported from 2,500 simulations for representative choices of 4 parameters: true heritability (h2), proportion of causal variants (r), prevalence (P) (NA for continuous phenotypes), and proportion of heritability from uncommon variants (α).
| h2 | r | P | α |
|
ĥ2 | |
|---|---|---|---|---|---|---|
| 0.8 | 0.01 | NA | 0.0 | 0.797(0.001) | 0.800(0.004) | |
| 0.8 | 0.001 | NA | 0.0 | 0.801(0.002) | 0.793(0.005) | |
| 0.5 | 0.01 | NA | 0.0 | 0.499(0.001) | 0.498(0.003) | |
| 0.5 | 0.001 | NA | 0.0 | 0.499(0.001) | 0.501(0.004) | |
| 0.8 | 0.01 | NA | 0.25 | 0.689(0.003) | 0.802(0.004) | |
| 0.8 | 0.01 | 0.2 | 0.25 | 0.691(0.002) | 0.782(0.005) | |
| 0.8 | 0.01 | 0.5 | 0.25 | 0.703(0.003) | 0.800(0.006) | |
| 0.8 | 0.01 | NA | 0.50 | 0.625(0.002) | 0.805(0.005) | |
| 0.8 | 0.01 | 0.5 | 0.50 | 0.637(0.003) | 0.797(0.006) | |
| 0.8 | 0.01 | NA | 1.0 | 0.473(0.002) | 0.796(0.005) | |
| 0.8 | 0.01 | 0.5 | 1.0 | 0.498(0.003) | 0.792(0.007) |
Setting individuals with the lowest P% of phenotypes as cases and all other as controls generated dichotomous phenotypes with prevalence P. The small number of individuals prevented simulation of case-control ascertainment, which may produce downward bias for low prevalence diseases in very large studies (see Supplementary Table 9 in [19]). Those biases are expected to be small in the prostate cancer data analyzed here because of the high prevalence of prostate cancer and moderate sample size. For large sample sizes, replacing mixed model based estimates with Haseman-Elston regression estimates will alleviate the issue of ascertainment bias20.
The results in Table 2 also demonstrate that complexities such as genotyping error, LD, or errors in local ancestry inference in African Americans do not introduce bias into the heritability estimates when phenotypes are generated under a non-infinitesimal mixture model. This may not be the case for other admixed populations such as Latinos21 (see Discussion).
Application to WHI, CARe, and AAPC cohorts
We applied our method to 21,497 African-American individuals from the WHI, CARe, and AAPC cohorts over a total of 12 quantitative phenotypes and 1 case-control phenotype (see Online Methods). Local ancestry was inferred using the HAPMIX, SABER+, and RFMix methods, which are extremely accurate in African Americans (r2=0.98 or greater)22,23,24. For each phenotype we estimated hg2, and by extension h2. For hg2 and we used the GCTA software package applied to the genotypes and local ancestry at each SNP respectively17. For those phenotypes measured in both cohorts we compute the inverse variance-weighted mean and standard error. For each phenotype we also list previously published estimates of heritability from family studies using twins and African-American estimates where available ( ) The results are shown in Table 3, and published African-American estimates are marked for reference. Estimates from European populations may not be directly comparable if the genetic or environmental bases for the phenotype differ substantially.
Table 3.
Heritability estimates of phenotypes from 21,497 African Americans from the WHI, CARe, and AAPC cohorts. Meta shows the inverse variance weighted meta-analysis for those phenotypes contained in both WHI and CARe data sets. is the previously published estimates of and is the previously published estimates of h2 from family studies. Published heritability studies of African Americans are denoted with a *.
| The heritability explained by genotyped SNPs ( ) in WHI and CARe. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype | WHI | s.e. | CARe | s.e. | Meta | s.e. |
|
|
| height | 0.461 | 0.058 | 0.378 | 0.029 | 0.395 | 0.026 | 0.4510 | |
| BMI | 0.198 | 0.055 | 0.078 | 0.065 | 0.148 | 0.042 | 0.1438 | |
| Log(HDL) | 0.316 | 0.057 | 0.224 | 0.066 | 0.277 | 0.043 | 0.1238 | |
| LDL | 0.294 | 0.056 | 0.156 | 0.067 | 0.238 | 0.043 | 0.1011 | |
| WBC | 0.725 | 0.051 | 0.848 | 0.091 | 0.755 | 0.044 | 0.211 | |
| WBC|FY | 0.188 | 0.054 | 0.167 | 0.097 | 0.183 | 0.047 | NA | |
| Log(TG) | 0.226 | 0.056 | NA | NA | NA | NA | NA | |
| Glucose | 0.16 | 0.063 | NA | NA | NA | NA | 0.1038 | |
| log(Insulin) | 0.086 | 0.051 | NA | NA | NA | NA | 0.0938 | |
| QT-interval | 0.251 | 0.098 | NA | NA | NA | NA | NA | |
| Log(CRP) | 0.295 | 0.056 | NA | NA | NA | NA | NA | |
| DBP | 0.148 | 0.053 | 0.170 | 0.066 | 0.157 | 0.041 | NA | |
| SBP | 0.162 | 0.054 | 0.189 | 0.066 | 0.173 | 0.042 | 0.2438 | |
| The total narrow sense heritability (ĥ2) as derived from in WHI and CARe. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype | WHI ĥ2 | s.e. | CARe ĥ2 | s.e. | Meta ĥ2 | s.e. |
|
|
| height | 0.611 | 0.135 | 0.503 | 0.120 | 0.550 | 0.090 | 0.7739* | |
| BMI | 0.252 | 0.097 | 0.208 | 0.085 | 0.227 | 0.064 | 0.4739* | |
| Log(HDL) | 0.418 | 0.117 | 0.470 | 0.146 | 0.438 | 0.091 | 0.5239* | |
| LDL | 0.395 | 0.116 | 0.333 | 0.140 | 0.370 | 0.089 | 0.5339* | |
| WBC | 3.267 | 0.322 | 3.703 | 0.447 | 3.415 | 0.261 | 0.4840* | |
| WBC|FY | 0.172 | 0.084 | 0.247 | 0.166 | 0.187 | 0.075 | NA | |
| Log(TG) | 0.225 | 0.094 | NA | NA | NA | NA | 0.4039* | |
| Glucose | 0.104 | 0.087 | NA | NA | NA | NA | 0.2941* | |
| log(Insulin) | 0.105 | 0.077 | NA | NA | NA | NA | 0.2841* | |
| QT-Interval | 0.336 | 0.164 | NA | NA | NA | NA | 0.4142* | |
| Log(CRP) | 0.542 | 0.139 | NA | NA | NA | NA | 0.5643* | |
| DBP | 0.179 | 0.088 | 0.238 | 0.119 | 0.200 | 0.071 | 0.1339* | |
| SBP | 0.187 | 0.092 | 0.233 | 0.117 | 0.205 | 0.072 | 0.1739* | |
| The complete AAPC results. | |||||||
|---|---|---|---|---|---|---|---|
| Phenotype | AAPC | s.e. |
|
AAPC ĥ2 | s.e. | ||
| PC | 0.182 | 0.040 | NA | 0.328 | 0.093 | 0.5844 | |
| PC|8q24 | 0.174 | 0.040 | NA | 0.315 | 0.092 | NA | |
Several phenotypes, including height, BMI, HDL, TG, PC, and WBC (conditioned on ancestry at the Duffy antigen locus FY; see below), had h2 estimates lower than family-based estimates. This could be due to the phenotype-specific effects of epistasis, gene environment interaction, and/or shared environmental factors that can inflate family based estimates12,13. In our recent work using an extended genealogy inclusive of more distantly related individuals we also found height and BMI estimates lower than previous heritability estimates, providing further evidence of inflation11. The lower estimates could also reflect a difference in the heritability between African Americans and the previous study populations. There were no statistically significant differences in h2 estimates between the cohorts.
Yang et al. proposed an adjustment to account for the incomplete coverage of genotyping platforms10. We applied this approach in the CARe data (see Supplementary Table 3), and observed an increase in hg2 of less than 1% in all phenotypes. We include genome-wide ancestry proportion as a fixed effect in our mixed model. If there exists an environmental factor that affects phenotype and is correlated with ancestry, our heritability estimates will discount this environmental effect leading to higher estimates of heritability. Specifically, it will remove the variance of the environmental factor that can be explained by ancestry from the environmental component ( ) in the denominator of the heritability estimate (see Online Methods).
Differences between our heritability estimates and those of previous studies can also be due to differences between the value of FSTC we used in this study and the true value of FSTC for the phenotype in question. Based on recent evidence that rare variants unlikely to contribute to a large proportion of phenotypic variation25,26, we computed an FSTC of 0.182 over the common variants (MAF > 5%) in African-Americans. However, this estimate drops to 0.165 for low-frequency variants (MAF < 5%) and 0.054 for rare variants (MAF < 1%). Estimates of heritability assuming a rare variant only phenotype model would be more than three times as large as from a common variant only phenotype model. Therefore, if rare variants contribute substantially to phenotypic variation or if balancing or negative selection constrained the genetic distance at causal variants, then our estimates of heritability will be biased downward (see Discussion).
Positive selection acting at causal variants could induce such a bias in FSTC, and we included WBC as a positive control for this type of bias. A SNP in the DARC gene (FY, Duffy null allele) is highly differentiated between CEU and YRI, likely due to its protective effect against vivax malaria27. It is also a SNP of large effect size for WBC28. Therefore, the average FSTC at causal variants for WBC, is much higher than the value 0.182 estimated from common variants (see Online Methods). The h2 estimate of WBC is 3.42 due to the effect of this positive selective pressure. Ancestry at the FY locus accounts for ∼20% of the phenotypic variation in WBC28. By including ancestry at FY as a fixed effect (WBC|FY) we obtain an h2 estimate of 0.19, which is lower than the published estimate of 0.48.
We perform a sensitivity analysis to assess whether this type of bias is likely to be problematic. Since strong positive selection is unusual29, we consider a single locus under positive selection. We estimate bias as a function of FSTC at the locus and the variance explained by the locus. The results in Supplementary Table 4 show that only for extreme values of both locus FSTC and heritability will there be significant bias in heritability due to positive selection. As an example we consider the 8q24 locus in prostate cancer, which contains causal SNPs that are highly differentiated SNPs between African and European ancestors, producing an admixture-mapping peak30. However, because this locus explains less than 2% of the heritability of prostate cancer, even exceedingly strong population differentiation at this locus will not substantially bias our overall results.
Partitioning heritability across the genome
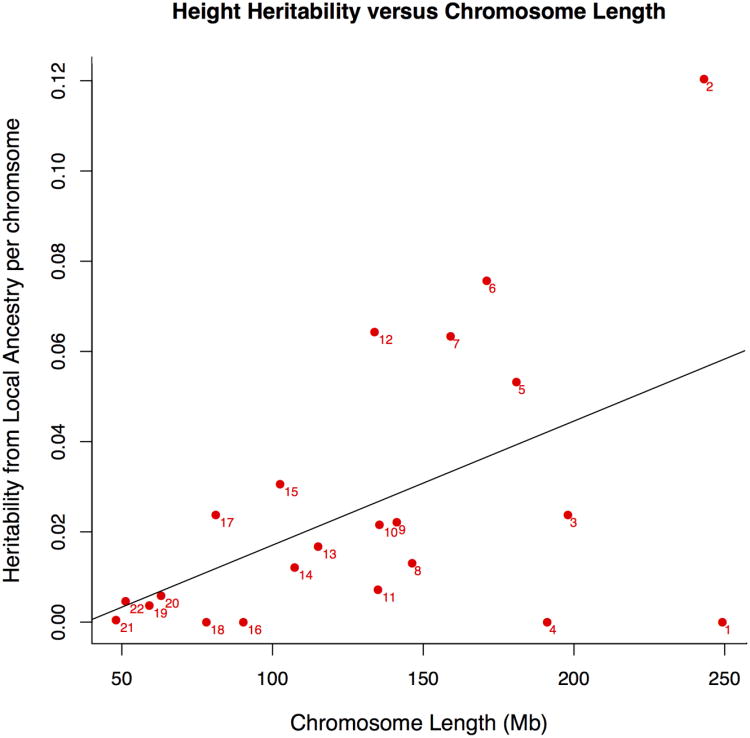
Our method is also capable of estimating the total narrow sense heritability attributable to a particular genomic region. This is accomplished by constructing the kinship matrix using just those ancestry segments in the region of interest and applying the variance component model to the phenotype of interest using the region-specific kinship matrix (see Online Methods). We partitioned the heritability for each of the phenotypes from the CARe data set across each of the chromosomes31. We applied weighted linear regression to determine the relationship between heritability and chromosome length (see Online Methods). The results for height are presented in Figure 2 and the full results are provided in Supplementary Table 2. We find a strong correlation between chromosome length and the heritability of height (Pearson correlation = 0.513, weighted p-value = 0.0028). logHDL, BMI, and SBP, also produced significant results (weighted p-value < 0.03, 0.02, 0.02 respectively). Other phenotypes had standard errors too large to produce meaningful results. To address this, we averaged the heritability from each chromosome across all phenotypes (using WBC|FY instead of WBC) and we observed a significant correlation between chromosome length and mean chromosomal heritability (Pearson correlation=0.686, weighted p-value <0.0002).
Figure 2.
Estimated heritability of height for each chromosome in the CARe data set. The numbers adjacent to each point are the chromosomes. We plot the regression line of h2 per chromosome regressed on chromosome length. We find a strong correlation between chromosome length and height heritability (Pearson correlation = 0.513, weighted p-value = 0.0028).
Discussion
We developed a method for estimating narrow sense heritability from unrelated individuals by leveraging the two ancestral genomes in recently admixed populations, such as African Americans. We used a population genetic approach to derive the relationship between heritability and variation in local ancestry in admixed populations. Theory and simulations confirm that under an infinitesimal phenotypic model our approach produces unbiased estimates of heritability. Since the individuals are distantly related, our approach will not produce heritability estimates inflated by epistasis, gene environment interactions, or shared environmental effects.
Our method is also able to partition total narrow-sense heritability (h2) along genomic segments such as chromosomes, as we have shown by application to the phenotypes in the CARe data set. This is distinct from recent work that instead partitioned the heritability explained by genotyped SNPs (hg2) across chromosomes31-33. While a previous method has also partitioned h2 along chromosomes34,35, it relies on the use of siblings, leading to very large standard errors, and is limited by the coarseness of shared IBD segments (which extend for tens of megabases). Our approach is limited by the coarseness of local ancestry segments (which extend for megabases) and thus cannot be applied at the level of individual genes.
We applied our method to an African-American population in this study. Application to more complex admixed populations such as Latinos will have to account for the reduced accuracy in local ancestry inference21 to avoid downward bias. Restricting to two ancestry categories (e.g. Native American vs. non-Native American ancestry)36 is one approach to handle multi-way admixture, but it may be possible to extend our derivation to multi-way admixture. There is evidence that African Americans have a small proportion of admixture from Native American populations (0.5%)24, but this very small proportion is unlikely to significantly change our results. Substantial errors in the assumed population genetic structure would perturb the values of FSTC and θ, and resulting h2 estimates would be biased in proportion to these errors. Application to sex chromosomes can be adapted from the approach taken in[31], but must be analyzed separately due to the differences in admixture proportion of European ancestry on autosomes and sex chromosomes.
In our previous work we found that heritability estimates from related individuals followed a pattern consistent with biases due to shared environment11. In this work we found that a linear additive model, implicitly including both rare and common variants, typically explained less phenotypic variation than that predicted in family studies. These new estimates of narrow-sense heritability are less susceptible to bias and provide additional evidence that family based estimates are inflated. Unlike [11], we were able to obtain estimates for both quantitative and case-control traits. We also found that chip based additive models explained less phenotypic variation than our estimates. In the meta-analyzed phenotypes common to CARe and WHI the average of these estimates were 24.7% and 31.1% respectively. Rare variants and poorly tagged common variants are the most likely explanation for the difference between these two estimates. We discuss other possible explanations below.
Our method does produce biased estimates when model assumptions are violated. Specifically, if the genetic distance we estimated over common variants (0.182; see Online Methods) differs from the distribution over causal variants, our method can be either inflated or deflated. If selection were acting on the causal variants their FSTC could be higher or lower depending on the direction of selection. In the case of positive selection in one of the ancestral groups but not the other, the true value of FSTC will be larger than our genome-wide estimate and so our h2 estimate will be inflated. For example, estimates for white blood cell count were larger than , due to strong selective pressure at the Duffy locus27,37. However, strong positive selection is believed to be rare in recent human evolution29. If a large proportion of phenotypic variance is due to rare variants then incorrect estimates of FSTC may induce bias. However, previous reports suggest that rare variation explains a small proportion of total heritability25,26.
The application of our approach to two large cohorts of African Americans revealed a difference between previously published family-based estimates of the heritability of height and BMI and our estimates. This suggests that there is a significant contribution of non-additive genetic effects or shared environmental effects that differ between MZ and DZ twins. The future application of our method to large-scale studies of African Americans will both provide a mechanism of estimating the total narrow sense heritability of phenotypes as well as determining the genetic architecture of complex phenotypes.
Online Methods
Given a set of M admixed individuals with two ancestral populations (P0 and P1), let the local ancestry for individual i at SNP s, γis ∊ {0,1,2}, be the number alleles inherited from a P1 ancestor. We use a mixed model approach to estimate the contribution of variation in local ancestry to phenotypic variation for the phenotype Y=y1, y2, …yM. We first construct a local ancestry based kinship matrix Kγ, which is constructed similarly to the genotyped-based kinship matrix K, but with local ancestry substituted for genotypes at each SNP. We then find the parameters and which maximize the likelihood of the mixed model . The heritability explained by local ancestry is given by h2g. Finally, we use the formula to estimate h2.
Definition of h2
Heritability is the ratio of genetic variance to the sum of genetic and environmental variance . In this case we are defining these elements with respect to an admixed population. For a given phenotype, both and can vary between the ancestral European and African populations. For example, will vary with ancestry if the minor allele frequency at causal variants is systematically larger in one of the two populations. It is also possible for ancestry to be associated with environmental factors. In this case, by conditioning on genome-wide ancestry, our method will remove the environmental variance that can be explained by ancestry and estimate the heritability of the component of phenotype that cannot be predicted by genome-wide ancestry, thereby increasing the heritability estimate.
Estimation of
We use a variance components approach to determine the phenotypic variance described by local ancestry using θ as a fixed effect to prevent confounding from environmental factors association with ancestry. This method is equivalent to recent methods used to determine the phenotypic variance described by genotyped SNPs ( ), replacing genotypes with inferred local ancestry10.
Derivation of relationship between h2 and
Let i denote (diploid) individuals and s index SNPs. Individual i is assigned global ancestry proportion θi from some distribution F(.) with mean E[θi] = θ and variance σ2θ. Given θi, an individual is assigned maternal and paternal local ancestries γi,s,M and γi,s,P at each SNP (0 or 1 copies of European ancestry), from Bernoulli distribution Ber(θi). Given local ancestries γi,s,M, γi,s,P and allele frequencies ps,o, ps,1 at SNP s in populations 0 and 1, individuals are assigned maternal genotypes gi,s,M =γi,s,M Zi,s,1 + (1-γi,s,M) Zi,s,0 where Zi,s,0∼Ber(ps,0) and Zi,s,1∼Ber(ps,1), and similarly for paternal genotypes. The diploid genotype gi,s= gi,s,P+ gi,s,M (0, 1 or 2), and the diploid local ancestry γi,s = γi,s,P+γi,s,M (0, 1 or 2).
We define E[gi,s] = μg,s and Var[gi,s] = σ2g,s, and the normalized genotype , where
| (1) |
| (2) |
Similarly, we define E[γi,s] = μγ and Var[γi,s] = σ2γ, and the normalized local ancestry at each locus , where
| (3) |
| (4) |
Although though equation (4) may not be strictly true (e.g. in a population where all individuals have 1 European parent and 1 African parent), it is approximately true for African Americans22. Furthermore, can be estimated empirically, and we do so in this work. We model the phenotype of individual i as
| (5) |
where , Var[yi]=1, E[yi]=0, the effect size of SNP s is βs, and . By substitution and algebra we get
| (6) |
Plugging into equation (5), we get
| (7) |
Note that δi does not depend on local ancestry, which allows us to compute the heritability due to local ancestry h2γ as:
| (8) |
We define FSTC as a measure genetic distance between ancestral populations weighted by the square of effect size βs:
| (9) |
This results in a final relationship
| (10) |
In practice we do not know the effect size of every SNP and must make simplifying assumptions about their distribution in order to estimate FSTC. First consider a simple phenotypic model in which genotypic effect size βs is independent of ps,o and ps,1. Then
| (11) |
where N is the number of SNPs. Then equation (8) becomes
| (12) |
The FSTC in equation (12) is a genome-wide measure of genetic difference between the ancestral populations. This is related to the classic parameter FST when all variants are causal (i.e. the infinitesimal model).
| (13) |
Now consider a more complex model in which the effect size of SNPs can fall into one of L classes such that the effect size distribution is a function of the class L. These classes could be, for example, rare and common variants (used in this work). We defined the genetic distance between ancestral populations within each class as FSTL and the phenotypic variance explained by SNPs in this class as h2L. Again substituting into equation (8) we have,
| (14) |
Therefore a weighted measure of genetic distance in each class.
To obtain an estimate of h2 we must estimate θ, FSTC, and . The parameter θ is estimated from local ancestry inference. The parameter FSTC is estimated from assumptions about the variance explained by SNPs in each genotypic class combined with external reference panels45,46.
Definition and Estimation of FSTC
As shown in the equations above we are defining FST to be the weighted average (across all SNPs s) of ratios . While this is similar to standard versions of FST, a ratio of averages is recommended instead when the goal is to draw population genetic inferences47. If the distribution of SNPs effect sizes is not a function FST then this would be the appropriate definition for our heritability estimation approach. However, recent work has shown that rare variants are unlikely to contribute to a large proportion of phenotypic variation48,25. As has been reported previously47, the average of ratios estimate will shrink when including many rare variants in the estimate. This is reflected in the 1000 Genomes based estimate of FST =0.07, which used an average of ratios49. Therefore, FST will produce a biased estimate of heritability because for the variance explained by rare variants is different from the variance explained by common variants. To account for this we defined a parameter FSTC, which is a weighted measure of genetic distance between ancestral populations (equation 9).
In practice we defined FSTC as the average FST within each class L of SNPs (FSTL), weighted by the proportion of phenotypic variance explained by that class:
| (15) |
Consider a situation in which L contains two classes, rare and common SNPs, with FST 0.054 and 0.182 respectively. If rare variants explained 10% of the heritability and common variants explain 90% of the heritability, then FSTC=0.1692. We estimated FSTC over the HapMap337 data set by using CEU and YRI as proxies for the ancestral populations of African-Americans, using an admixture proportion of 18.3% European ancestry, and assuming distribution of causal variant frequencies. We estimated a value of 0.182 assuming causal variant MAF > 5% (which we used in this work), 0.165 assuming MAF < 5%, and 0.054 assuming MAF < 1%.
Simulations with Simulated Genotypes
In order to examine the properties of our approach, we first applied our method to data generated under a simple simulation framework for generating genotypes, local ancestries, and phenotypes of individuals from an admixed population. Allele frequencies pA1, pA2, …,pAN of N SNPs from an ancestral population were drawn uniformly from [0.1-0.9]. Allele frequencies of SNPs from P0 were drawn from a beta distribution with parameters p(1- FSTC)/FSTC and (1-p)(1- FSTC)/FSTC for each SNP s, and similarly for P1. The parameter FSTC determines the genetic distance between the two populations. The global proportion of P0 ancestry θ1, θ2, …θM for each of M individuals was drawn either uniformly from [0.4,0.6], from the normal distribution N(0.5,0.1), or fixed at 0.5. Local ancestry for individual i at SNP s (γis), was generated by two draws from binomial distribution with parameter θs. The genotypes from individual i at SNP s(gis) were then generated by drawing from the binomial distributions with allele frequencies specified by the local ancestry for that individual at that SNP. That is, if the individual had two copies of ancestry from P0 at SNP s then two draws from a binomial with parameter p0s were used. To create a phenotype we first selected Nr causal variants where r is the proportion of causal variants. Effect sizes were drawn from the normal distribution N(0,h2/(Nr)) and the genetic element of the phenotype was generated by taking the inner product of the causal variants, normalized to have mean 0 and variance 1, and the effect sizes for the variants. Normally distributed random noise was added such that the total heritability in the population was h2.
Simulations with Real Genotypes
We split the genotypes from 5,129 distantly related CARe individuals into two groups. The common group contained those SNPs with MAF > 5% in both CEU and YRI. The uncommon group contained all other SNPs (i.e. MAF < 5% in either or both of CEU and YRI). The genotype kinship matrix K was constructed over the common SNPs and the local ancestry kinship matrix Kγ was constructed using the local ancestry called at every 5th common SNP.
We simulated a phenotype by first selecting a proportion r of causal variants at random from the common and uncommon SNPs, leaving Nc common causal and Nn uncommon causal SNPs. We then selected a fraction of phenotypic variance α explained by the uncommon SNPs. At α=0.0 uncommon variants had no effect and the genetic basis of the phenotype was entirely determined by common variants. We then chose effect sizes for each common and uncommon SNP by drawing from normal distributions N(0,(1-α)h2/(Nc)) and N(0,(α)h2/(Nn)) respectively. The genetic element of the phenotype was generated by taking the inner product of the causal variants, normalized to have mean 0 and variance 1 in the admixed population, and the effect sizes for the variants. Normally distributed random noise was added such that the total heritability in the population was h2. The FSTC of the common and uncommon SNPs was 0.15 and 0.25 respectively. The study FSTC used to estimate heritability was the weighted mean 0.15(1-α) + 0.25α as described in the derivation above. Setting individuals with the lowest P% of phenotypes as cases and all other as controls generated dichotomous phenotypes with prevalence P.
Data set approvals
The CARe project has been approved by the Committee on the Use of Humans as Experimental Subjects (COUHES) of the Massachusetts Institute of Technology, and by the Institutional Review Boards of each of the nine parent cohorts.
The WHI project has been approved by the Human Subjects Committees at the WHI Clinical Coordinating Center (FHCRC) and at the 40 WHI Field Centers.
The AAPC project has been approved by the Institutional Review Board of the University of Southern California. The 11 studies contributing to the AAPC each received approval for the use of specimens from their patients.
CARe data set
Affymetrix 6.0 genotyping and QC filtering of African-American samples from the CARe cardiovascular consortium was performed as described previously50. After QC filtering for each of ARIC, CARDIA, CFS, JHS and MESA cohorts and subsequent merging, 8,367 samples and 770,390 SNPs remained. To limit relatedness among samples we restricted all analyses to a subset of 5,129 samples in which all pairs have genome-wide relatedness of 0.05 or less and had between 5% and 45% European ancestry. We performed local ancestry inference using the HAPMIX software with the CEU and YRI HapMap populations as reference ancestral populations. We examined seven phenotypes from the CARe cohort, height, body mass index (BMI), log transformed high density lipoprotein cholesterol (logHDL), low density lipoprotein cholesterol (LDL), white blood cell count (WBC), diastolic blood pressure (DBP), and systolic blood pressure (SBP). For each phenotype we included age, sex, study center, proportion of European ancestry, and the top 5 principal components as fixed effects. A detailed description of the phenotypes can be found here51.
WHI Data Set
Affymetrix 6.0 genotyping and QC filtering of African-American samples from the Women's Health Initiative (WHI) SNP Health Association Resource (SHARe) was performed as described previously52. The dataset includes extensive phenotypic and genotypic data on 12,008 African American and Hispanic women aged 50-79 enrolled in one or more components of the WHI program. We included only African American samples and to limit relatedness among samples we restricted all analyses to a subset of 8,153 samples in which all pairs have genome-wide relatedness of 0.05 or less. We performed local ancestry inference using the SABER+23 software with the CEU and YRI HapMap populations as reference ancestral populations. We examined 10 phenotypes from the WHI cohort (BMI), log transformed high density lipoprotein cholesterol (logHDL), low density lipoprotein cholesterol (LDL), white blood cell count (WBC), log transformed triglycerides (logTG), glucose, log transformed insulin (logInsulin), QT interval duration (QT-INTERVAL), C-reactive protein (CRP), diastolic blood pressure (DBP), and systolic blood pressure (SBP). For each phenotype we included age and proportion of European ancestry as fixed effects. A detailed description of the phenotypes can be found here52.
African American Prostate Cancer Data Set (AAPC)
IlluminaHuman1M-Duov3_B genotyping and QC filtering of African-American samples from the African American Prostate Cancer Study (AAPC) from a total of 11 participating studies was performed as described previously55,53,54. The cleaned dataset includes 9,641 African American subjects and 1,001,899 autosomal SNPs. To limit relatedness among samples we restricted all analyses to a subset of 8,215 samples in which all pairs have genome-wide relatedness of 0.05 or less. We performed local ancestry inference using the RFMix24 with the CEU and YRI HapMap populations as reference ancestral populations. We examined prostate cancer (PC) outcome for each subject. There were 4207 cases and 4008 controls after QC. Due to the admixture signal at the 8q24 locus54, we also estimated heritability removing 8q24 from the SNPs used to estimate the kinship (PC|8q24). For each phenotype we included age and the top 10 principal components as fixed effects. For conversion to the liability scale we used a prevalence of 5%54.
Partitioning Heritability across the genome
To estimate the heritability for a particular genomic segment we compute the genetic relatedness matrix as defined in Yang et al10, replacing genotypic for local ancestry calls, and restricting to just those SNPs contained in the region of interest. Given a partitioning of segments along the genome (in our case 22 segments), it is possible to fit them individually or jointly. We attempted both approaches, but found that the joint fit produced a numerical instability in the optimization algorithm preventing convergence. Thus all results reported for the single chromosome analyses are provided by individual and not joint estimates.
We performed both weighted and standard linear regression to assess the relationship between the heritability explained by a chromosome and the length of the chromosome. The weighted version accounts for the differences in number of SNPs contained in longer and shorter chromosomes and the weighting factor we used was the length of the chromosome in centimorgans.
Supplementary Material
Table 4.
Number of individuals for each phenotype in the CARe and WHI data sets. The AAPC data set contained 4207 PC cases and 4008 controls.
| Phenotype | WHI | CARe |
|---|---|---|
| height | 8109 | 5024 |
| BMI | 8153 | 5026 |
| Log(HDL) | 8014 | 4928 |
| LDL | 7979 | 4794 |
| WBC | 8035 | 3367 |
| WBC|FY | 8035 | 3367 |
| Log(TG) | 8015 | NA |
| Glucose | 6826 | NA |
| log(Insulin) | 7749 | NA |
| QT-Interval | 4143 | NA |
| Log(CRP) | 8014 | NA |
| DBP | 8153 | 5030 |
| SBP | 8153 | 5029 |
Acknowledgments
This research was supported by NIH grants R01 HG006399, R01 GM073059, and, R21 ES020754. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, HHSN271201100004C.
Footnotes
Author Contributions: N.Z., B.P., S.S., G.B., A.G., B.J.V.,C.H., J.G.W., C.K., D.S., A.P.R., H.T., and A.L.P. designed experiments.N.Z., J.Z., T.Y., A.T., S.P., H.T., and A.L.P performed experiments. N.Z., S.S., C.H., J.G.W., C.K., D.S., A.P.R., H.T., and A.L.P wrote text. T.L.A, S.I.B, W.J.B., S.C., N.F., P.G.G., J.H., A.JM.H., A.H., S.A.I., W.I., R.A.K., E.A.K., L.A.L, B.N., N.P., D.R., B.A.R., J.L.S., V.L.S., V.L.S., S.S.S., E.A.W., J.S.W., J.X. provided data.
References
- 1.Wray NR, et al. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–15. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaitlen N, Kraft P. Heritability in the genome-wide association era. Hum Genet. 2012 doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visscher PM, Brown MA, McCarthy MI, Yang J. Five Years of GWAS Discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee N, et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45:400–5. 405e1–3. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--concepts and misconceptions. Nat Rev Genet. 2008;9:255–66. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 8.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaitlen N, et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch M, Walsh B. Genetics and analysis of quantitative traits. xvi. Sinauer; Sunderland, Mass: 1998. p. 980. [Google Scholar]
- 14.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatia G, et al. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am J Hum Genet. 2011;89:368–81. doi: 10.1016/j.ajhg.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sham PC, Purcell S. Equivalence between Haseman-Elston and variance-components linkage analyses for sib pairs. Am J Hum Genet. 2001;68:1527–32. doi: 10.1086/320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating Missing Heritability for Disease from Genome-wide Association Studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–6. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golan D, Rosset S. Narrowing the gap on heritability of common disease by direct estimation in case-control GWAS. 2013;5363 [Google Scholar]
- 21.Pasaniuc B, et al. Analysis of Latino populations from GALA and MEC studies reveals genomic loci with biased local ancestry estimation. Bioinformatics. 2013;29:1407–1415. doi: 10.1093/bioinformatics/btt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson NA, et al. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet. 2011;7:e1002410. doi: 10.1371/journal.pgen.1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93:278–88. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46:220–4. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison AC, et al. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat Genet. 2013;45:899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamblin MT, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet. 2000;66:1669–79. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich D, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez RD, et al. Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–4. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–25. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SH, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–50. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visscher PM, et al. Genome partitioning of genetic variation for height from 11,214 sibling pairs. Am J Hum Genet. 2007;81:1104–10. doi: 10.1086/522934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemani G, et al. Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am J Hum Genet. 2013;93:865–75. doi: 10.1016/j.ajhg.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price AL, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–36. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nalls MA, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82:81–7. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vattikuti S, Guo J, Chow CC. Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits. PLoS Genet. 2012;8:e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson JG, et al. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis. 2005;15:S6-30–37. [PubMed] [Google Scholar]
- 40.Reiner AP, et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freedman BI, et al. Genome-wide scans for heritability of fasting serum insulin and glucose concentrations in hypertensive families. Diabetologia. 2005;48:661–8. doi: 10.1007/s00125-005-1679-5. [DOI] [PubMed] [Google Scholar]
- 42.Akylbekova EL, et al. Clinical correlates and heritability of QT interval duration in blacks: the Jackson Heart Study. Circ Arrhythm Electrophysiol. 2009;2:427–32. doi: 10.1161/CIRCEP.109.858894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox ER, et al. Epidemiology, heritability, and genetic linkage of C-reactive protein in African Americans (from the Jackson Heart Study) Am J Cardiol. 2008;102:835–41. doi: 10.1016/j.amjcard.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hjelmborg JB, et al. The Heritability of Prostate Cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol Biomarkers Prev. 2014 doi: 10.1158/1055-9965.EPI-13-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pennisi E. Genomics. 1000 Genomes Project gives new map of genetic diversity. Science. 2010;330:574–5. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
- 47.Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23:1514–21. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91:1011–21. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lettre G, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasaniuc B, et al. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium. PLoS Genet. 2011;7:e1001371. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franceschini N, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet. 2013;93:545–54. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolonel LN, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–57. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haiman CA, et al. Characterizing genetic risk at known prostate cancer susceptibility loci in African Americans. PLoS Genet. 2011;7:e1001387. doi: 10.1371/journal.pgen.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olama AA, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1101–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
