Abstract
Human dendritic cells (DCs) develop from progressively restricted bone marrow (BM) progenitors: these progenitor cells include granulocyte, monocyte and DC progenitor (GMDP) cells; monocyte and DC progenitor (MDP) cells; and common DC progenitor (CDP) and DC precursor (pre-DC) cells. These four DC progenitors can be defined on the basis of the expression of surface markers such as CD34 and hematopoietin receptors. In this protocol, we describe five multiparametric flow cytometry panels that can be used as a tool (i) to simultaneously detect or phenotype the four DC progenitors, (ii) to isolate DC progenitors to enable in vitro differentiation or (iii) to assess the in vitro differentiation and proliferation of DC progenitors. The entire procedure from isolation of cells to flow cytometry can be completed in 3–7 h. This protocol provides optimized antibody panels, as well as gating strategies, for immunostaining of BM and cord blood specimens to study human DC hematopoiesis in health, disease and vaccine settings.
INTRODUCTION
DCs are a heterogeneous population of immune cells that are essential mediators of immunity and tolerance1-3. Although DCs share a common ability to process and present antigen to naive T cells for the initiation of an immune response, they are not all identical, and their unique abilities and phenotypes are current areas of investigation. DCs from humans, which are best defined in the blood, are identified with BDCA markers. BDCA-2(CD303)+ plasmacytoid DCs (pDCs) have the unique ability to rapidly produce abundant type I interferon (IFN) in response to viral infection4. BDCA-1(CD1c)+ conventional DCs (cDCs) have been proposed to excel in CD4+ T-cell priming5,6. Finally, BDCA-3(CD141)hi cDCs have the ability to capture dead cells and to cross-present exogenous antigen, offering a mechanism for priming CD8+ T cells specific for pathogens that do not directly infect DCs7-12.
Whereas DC development has been studied extensively in mice13, the origin of human DCs and their relation to monocytes have been long debated. Our group has recently clarified the pathway for human DC hematopoiesis and shown its sequential origin from increasingly restricted but well-defined BM progenitors14,15 (Fig. 1). Human granulocyte, monocyte and DC lineages originate from a common progenitor, the GMDP. GMDPs develop into a more restricted human MDP. MDPs give rise to monocytes and a CDP, which loses the potential to produce monocytes and is restricted to produce the three major subsets of DCs. These committed DC progenitors reside in BM, as well as in cord blood (CB), but not in blood or lymphoid tissues14. Finally, CDPs give rise to pDCs, as well as to a circulating cDC precursor cell (pre-cDC). Indeed, pre-cDCs develop in the BM, travel through the blood and differentiate into the two subsets of cDCs in peripheral lymphoid organs. In studies of human volunteers injected with Fms-related tyrosine kinase 3 ligand (FLT3L), we showed that human pre-cDCs are mobilized into the blood similarly to the more differentiated DC subsets15. Now that the lineage-committed progenitors and immediate precursors for human DCs have been identified, studies to further define human DC hematopoiesis in health, disease and vaccine settings are possible, as well as the exploration of their potential utility in cellular immunotherapies. This can be facilitated by the use of this protocol, in which we describe flow cytometry assays to isolate and characterize DC progenitors.
Figure 1.
Schematic view of human dendritic cell (DC) hematopoiesis. DC hematopoiesis is initiated in the bone marrow (BM). A granulocyte, monocyte and DC progenitor (GMDP) develops into a monocyte and DC progenitor (MDP). MDPs give rise to monocytes and a common DC progenitor (CDP), which loses the potential to produce monocytes. CDPs give rise to plasmacytoid DCs (pDCs), as well as a circulating cDC precursor (pre-cDC). Pre-cDCs migrate from the BM through the blood to the periphery to produce the two major subsets of conventional DCs (cDCs)—i.e., BDCA-1+ cDCs and BDCA-3hi cDCs.
Development of the protocol
The study of human DC hematopoiesis has been hampered by the absence of validated markers to identify and track progenitors. Human hematopoietic stem and progenitor cells (HSPCs) are commonly defined by the expression of the cell surface protein CD34, as well as the non-expression of lineage antigens that are present on mature leukocytes. These Lin− (lineage) CD34+ HSPCs comprise only a small and variable fraction of human BM cells (2–4%), CB cells (~1%) and peripheral blood (PB; <0.2%). Human HSPCs have been further fractionated using common markers such as CD38, CD90, CD45RA, CD117 (stem cell factor (SCF) receptor) and CD135 (FLT3 receptor)16-19. However, the combination of these markers does not separate DC lineage–committed progenitors from multipotential progenitors. Moreover, it has been reported that a small fraction of cells that lack the expression of CD34 have progenitor potential20. These Lin− CD34− cells probably represent precursor cells that have lost CD34 expression but that are still too immature to express lineage markers. Therefore, CD34, as well as the afore-mentioned markers, are not enough to further separate HSPCs, and additional markers are required to identify and isolate DC lineage–committed progenitors.
As hematopoiesis relies on instructive cues from the BM niche such as hematopoietins, we hypothesized that the lineage potential of a progenitor might be determined by the set of growth factor receptors that it expresses. We showed that multipotential progenitors such as myeloid and lymphoid progenitors were heterogeneous, and they could be further fractionated using antibodies against receptors for granulocyte–macrophage colony-stimulating factor (GM-CSFR; CD116), macrophage colony-stimulating factor (M-CSFR; CD115) and interleukin-3 (IL-3R; CD123) (refs. 14,15). By using two slightly different antibody panels, we identified the following: (i) the human DC lineage–committed GMDP, MDP and CDP in the Lin− CD34+ fraction by combining antibodies against CD45RA, CD116, CD115 and CD123, as well as CD38 (ref. 14), and (ii) the human pre-cDC, which was found in the Lin− CD34− fraction and identified on the basis of the expression or lack of CD45RA, CD116, CD115 and CD123, as well as CD135 and CD117 (ref. 15). These progenitors are rare, which presents a challenge for the isolation of these cells. When working with scarce cells, the simultaneous analysis of all four DC progenitors seems to be essential in order to provide a comprehensive view of DC hematopoiesis.
The proven multicolor panels presented in this protocol are based on our original studies14,15 with modifications enabling simultaneous assessment of all DC lineage–committed progenitors. As the common feature of progenitors that are capable of developing into DCs is surface expression of CD117 and CD135, the panel to define them includes the following: (i) antibodies specific to CD117 and CD135, as well as CD34, CD123, CD115, CD116 and CD45RA; (ii) antibodies specific to CD3 (T cell), CD19 (B cell), CD14 (monocytes), CD335 (natural killer (NK) cell), CD66b (granulocytes) and CD10 (lymphoid progenitors) to exclude specific lineages (Lin−); (iii) antibodies specific to BDCA-1, BDCA-2 and BDCA-3 to exclude differentiated DCs (BDCA−); (iv) a viability dye to exclude dead cells; and (v) an optional antibody specific to CD45 to visualize hematopoietic cells in nonhematopoietic tissues or in vitro cultures. All antibodies (Table 1) and fluorochrome combinations (Table 2) are commercially available, and experiments can be run on commonly available flow cytometers and flow sorters that are capable of multicolor analysis (see configurations in Tables 3 and 4).
TABLE 1.
List of monoclonal antibodies used in this study.
| Marker | Fluorochrome | Clone | Host species and isotype | Manufacturer | Cat. no. |
|---|---|---|---|---|---|
| BDCA-2 (CD303) | FITC | AC144 | Mouse IgG1 | Miltenyi | 130-090-510 |
| CD116 | FITC | 4H1 | Mouse IgG1 | BioLegend | 305906 |
| BDCA-3 (CD141) | PE | AD5-14H12 | Mouse IgG1 | Miltenyi | 130-090-514 |
| CD135 | PE | 4G8 | Mouse IgG1 | BD Biosciences | 558996 |
| CD45 | PE Texas Red | HI30 | Mouse IgG1 | Life Technologies | MHCD4517 |
| HLA-DR | PE Texas Red | TÜ36 | Mouse IgG2b | Life Technologies | MHLDR17 |
| CD45RA | PE Texas Red | MEM-56 | Mouse IgG2b | Life Technologies | MHCD45RA17 |
| CD3 | PE Texas Red | S4.1 | Mouse IgG2a | Life Technologies | MHCD0317 |
| CD19 | PE Texas Red | SJ25-C1 | Mouse IgG1 | Life Technologies | MHCD1917 |
| CD14 | PE Texas Red | TüK4 | Mouse IgG2a | Life Technologies | MHCD1417 |
| BDCA-2 (CD303) | PerCP-Cy5.5 | 201A | Mouse IgG2a | BioLegend | 354210 |
| CD66b | PerCP-Cy5.5 | G10F5 | Mouse IgM | BioLegend | 305108 |
| CD335 | PerCP-Cy5.5 | 9E2 | Mouse IgG1 | BioLegend | 331920 |
| CD10 | PerCP-Cy5.5 | HI10a | Mouse IgG1 | BioLegend | 312216 |
| CD335 | PE-Cy7 | 9E2 | Mouse IgG1 | BD Biosciences | 562101 |
| BDCA-3 | PE-Cy7 | M80 | Mouse IgG1 | BioLegend | 344110 |
| HLA-DR | PE-Cy7 | L243 | Mouse IgG2a | BioLegend | 307616 |
| CD11c | PE-Cy7 | 3.9 | Mouse IgG1 | BioLegend | 301608 |
| CD33 | PE-Cy7 | WM53 | Mouse IgG1 | eBioscience | 25-0338-42 |
| CD38 | PE-Cy7 | HIT2 | Mouse IgG1 | BioLegend | 303516 |
| CD117 | Brilliant Violet 421 | 104D2 | Mouse IgG1 | BioLegend | 313216 |
| BDCA-3 (CD141) | Brilliant Violet 421 | M80 | Mouse IgG1 | BioLegend | 344114 |
| CD117 | Brilliant Violet 510 | 104D2 | Mouse IgG1 | BioLegend | 313220 |
| CD123 | Brilliant Violet 510 | 6H6 | Mouse IgG1 | BioLegend | 306022 |
| CD45 | V500 | HI30 | Mouse IgG1 | BD Biosciences | 560777 |
| CD10 | Brilliant Violet 605 | HI10a | Mouse IgG1 | BioLegend | 312222 |
| CD20 | Brilliant Violet 605 | 2H7 | Mouse IgG2b | BioLegend | 302333 |
| CD123 | Brilliant Violet 650 | 6H6 | Mouse IgG1 | BioLegend | 306019 |
| CD45RA | Qdot 655 | MEM-56 | Mouse IgG2b | Invitrogen | Q10069 |
| CD14 | Qdot 800 | TüK4 | Mouse IgG2a | Invitrogen | Q10064 |
| BDCA-2 | APC | AC144 | Mouse IgG1 | Miltenyi | 130-090-905 |
| CD115 | APC | 9-4D2-1E4 | Rat IgG1 | BioLegend | 347306 |
| Clec9A | Alexa Fluor 700 | 683409 | Mouse IgG1 | R&D Systems | FAB6049N |
| CD34 | Alexa Fluor 700 | 581 | Mouse IgG1 | BioLegend | 343526 |
| BDCA-1 (CD1c) | APC-Cy7 | L161 | Mouse IgG1 | BioLegend | 331520 |
| CD3 | BUV395 | UCHT1 | Mouse IgG1 | BD Biosciences | 563546 |
| CD19 | BUV395 | SJ25-C1 | Mouse IgG1 | BD Biosciences | 563549 |
Table 2.
Antibody panels for human dendritic cell progenitors.
| Panel | Panel #1, 15 color (identification) |
Panel #2, 15 color (characterization) |
Panel #3, 11 color (isolation) |
Panel #4, 13 color (culture output) |
Panel #5, 14 color (culture proliferation) |
|---|---|---|---|---|---|
| FITC | CD116 | CD116 | CD116 | BDCA-2 | CFSE |
| PerCP Cy5.5 | CD66b/CD335/BDCA-2 | CD66b/CD335/BDCA-2 | CD66b/CD335/BDCA-2/CD10 | CD66b | CD66b |
| PE | CD135 | CD135 | CD135 | BDCA-3 | BDCA-3 |
| PE Texas Red | CD45 | CD45RA | CD3/CD19/CD14 | HLA-DR | HLA-DR |
| PE-Cy7 | BDCA-3 | (antibody of choice) | BDCA-3 | CD335 | CD335 |
| APC | CD115 | CD115 | CD115 | — | BDCA-2 |
| Alexa Fluor 700 | CD34 | CD34 | CD34 | Clec9A | Clec9A |
| APC-Cy7 | BDCA-1 | BDCA-1 | BDCA-1 | BDCA-1 | BDCA-1 |
| BV421 | CD117 | BDCA-3 | CD117 | — | — |
| V500, BV510 | CD123 | CD117 | CD123 | CD45 | CD45 |
| BV605 | CD10 | CD10 | — | CD20 | CD20 |
| Qdot 655, BV650 | CD45RA | CD123 | CD45RA | CD123 | CD123 |
| Qdot 800 | CD14 | CD14 | — | CD14 | CD14 |
| Live/Dead | Live/Dead blue | Live/Dead blue | — | Live/Dead blue | Live/Dead blue |
| BUV395 | CD3/CD19 | CD3/CD19 | — | CD3 | CD3 |
Table 3.
Configuration of LSR II.
| Detector | Long-pass filter | Band-pass filter | Fluorochrome |
|---|---|---|---|
| Blue laser (488 nm) | |||
| A | 685LP | 695/40 | PerCP-Cy5.5 |
| B | 505LP | 530/30 | FITC, CFSE |
| C | 488/10 | SSC | |
| Yellow/green laser (561 nm) | |||
| A | 750LP | 780/60 | PE-Cy7 |
| D | 600LP | 610/20 | PE Texas Red |
| E | — | 582/15 | PE |
| Red laser (633 nm) | |||
| A | 735LP | 780/60 | APC-Cy7 |
| B | 690LP | 710/50 | Alexa Fluor 700 |
| C | — | 660/20 | APC |
| Violet laser (405 nm) | |||
| A | 635LP | 655/20 | Qdot 655, BV650 |
| B | 595LP | 605/20 | BV605 |
| C | 505LP | 525/50 | V500, BV510 |
| D | — | 450/50 | BV421 |
| UV laser (355 nm) | |||
| A | 735LP | 780/60 | Qdot 800 |
| B | 410LP | 450/50 | Live/Dead blue |
| C | — | 379/28 | BUV395 |
Table 4.
Configuration of ARIA III.
| Detector | Long-pass filter | Band-pass filter | Fluorochrome |
|---|---|---|---|
| Blue laser (488 nm) | |||
| A | 655LP | 695/40 | PerCP-Cy5.5 |
| B | 502LP | 530/30 | FITC |
| C | — | 488/10 | SSC |
| Yellow/green laser (561 nm) | |||
| A | 735LP | 780/60 | PE-Cy7 |
| D | 600LP | 610/20 | PE Texas Red |
| E | — | 582/15 | PE |
| Red laser (640 nm) | |||
| A | 735LP | 780/60 | APC-Cy7 |
| B | 690LP | 730/45 | Alexa Fluor 700 |
| C | — | 660/20 | APC |
| Violet laser (407 nm) | |||
| A | 600LP | 660/20 | Qdot 655 |
| B | 502LP | 510/50 | BV510 |
| C | — | 450/40 | BV421 |
Defining human DC progenitors also required that we develop an efficient culture assay to study the ability of the progenitors to proliferate and differentiate in response to hematopoietic growth factors. Through systematic testing of different combinations of cytokines, as well as the use of mouse stromal cell lines, we showed that the in vitro differentiation of CD34+ HSPCs on MS-5 stromal cells in addition to FLT3L, SCF and GM-CSF cytokines (referred to as MS5+FSG herein) support the development of multiple lineages, including all three major human DC subsets. The resulting DC subsets resemble their blood counterparts both in gene expression and function14.
Experimental design
In this protocol, we give a flow cytometry–based workflow for (i) simultaneous detection of DC progenitors (Table 2, panel #1 and Fig. 2), as well as their characterization (Table 2, panel #2 and Fig. 3); (ii) isolation of DC progenitors to enable in vitro differentiation (Table 2, panel #3 and Fig. 4); and (iii) evaluation of DC progenitor in vitro differentiation (Table 2, panel #4 and Fig. 5) and proliferation (Table 2, panel #5 and Fig. 6). The procedure also covers various preparatory steps—isolation, freezing and thawing of mononuclear cells (MNCs) and hematopoietic differentiation on stromal cells.
Figure 2.
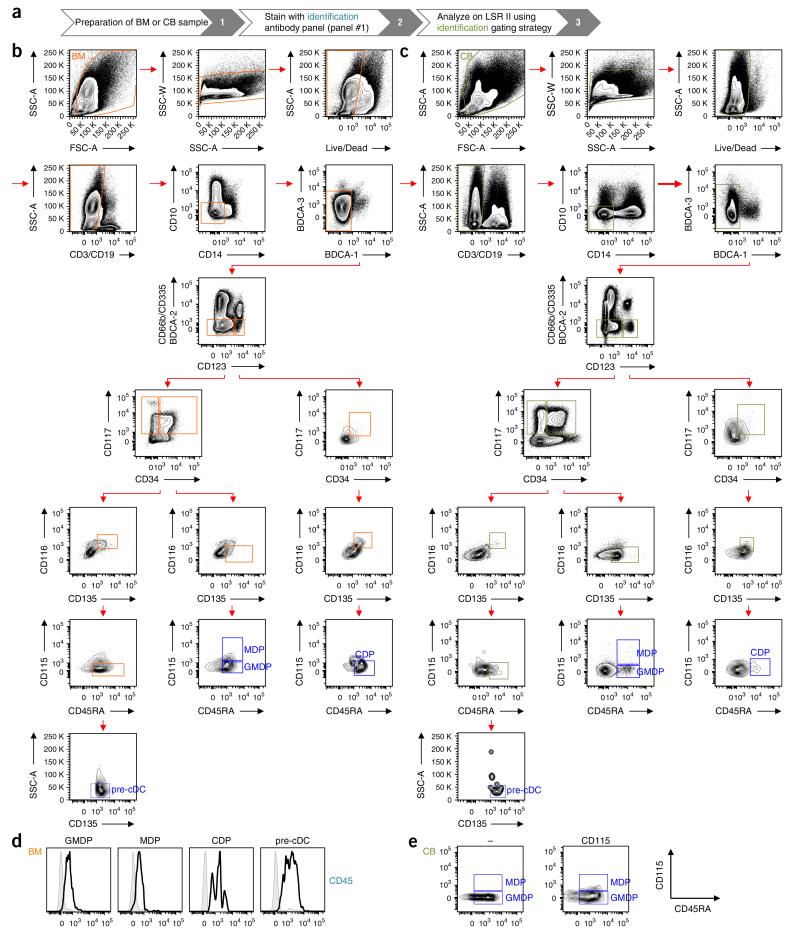
Gating strategy for the identification of human DC progenitors. (a) Workflow for the identification of DC progenitors. (b,c) Human BM (b) and CB (c) MNCs were stained as indicated in Table 2 (panel #1). Progenitors are identified by first gating on live cells (FSC-A versus SSC-A), and then on singlets (SSC-A versus SSC-W) and live cells (Live/Dead blue versus SSC-A). Lin− BDCA− cells are obtained by successively gating out CD3+, CD19+ cells (CD3, CD19 versus SSC-A), CD14+, CD10+ cells (CD14 versus CD10), BDCA-1+, BDCA-3hi cells (BDCA-1 versus BDCA-3) and CD66b+, CD335+, BDCA-2+ cells (CD66b, CD335, BDCA-2 versus CD123). Lin−BDCA−CD123−/lo and Lin−BDCA−CD123hi populations are further subgated on the basis of the expression of CD34 versus CD117, CD135 versus CD116 and CD45RA versus CD115. In these gated populations, GMDPs are CD123−/loCD34+CD117+CD135+CD116−CD115−CD45RA+; MDPs are CD123−/loCD34+CD117+CD135+CD116−CD115+CD45RA+; CDPs are CD123hiCD34+CD117+CD135+CD116+CD115−CD45RA+; and pre-cDCs are CD123−/lo D34−CD117+CD135+CD116+CD115−CD45RA+SSC-Alo. FSC, forward scatter; SSC, side scatter. (d) Expression of CD45 by GMDPs, MDPs, CDPs and pre-cDCs from human BM. Histograms show the expression of stained (black line) and unstained (gray line) cells. (e) FMO control for CD115 staining used to discriminate between GMPs and MDPs in CB.
Figure 3.
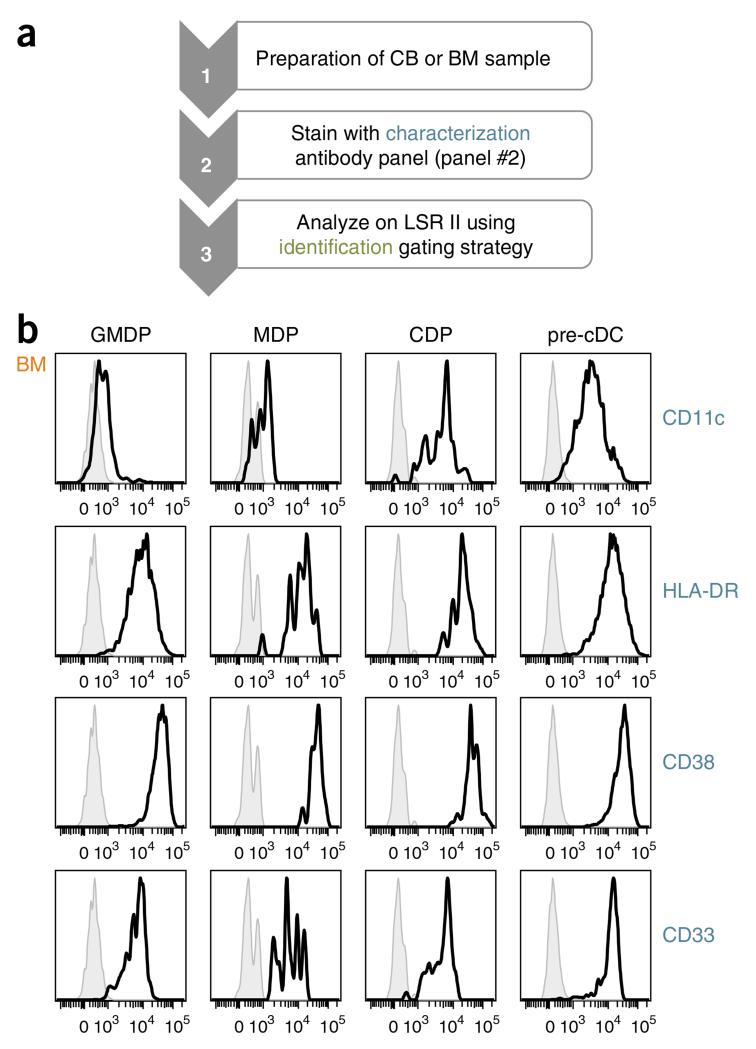
Characterization of human DC progenitors. (a) Workflow for the characterization of DC progenitors. (b) Expression of CD11c, HLA-DR, CD38 and CD33 by GMDPs, MDPs, CDPs and pre-cDCs from human BM. DC progenitors are defined as in Figure 2b. Histograms show expression (x axis) and number of cells (y axis) of stained (black line) and unstained (gray line) cells.
Figure 4.
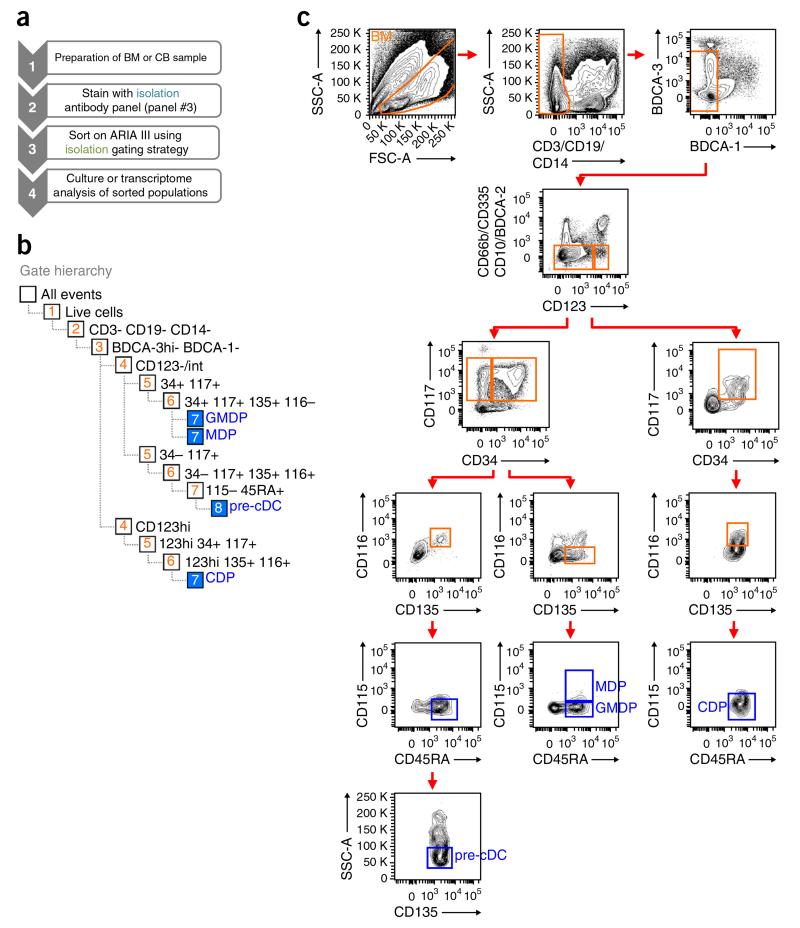
Gating strategy for the isolation of human DC progenitors. (a) Workflow for the isolation of DC progenitors. (b) Gate hierarchy for sorting. (c) Human BM MNCs were stained as indicated in Table 2 (panel #3). Progenitors are identified by first gating on live cells (FSC-A versus SSC-A). Lin− BDCA− cells are obtained by successively gating out CD3+, CD19+, CD14+ cells (CD3, CD19 and CD14 versus SSC-A), BDCA-1+, BDCA-3hi cells (BDCA-1 versus BDCA-3) and CD66b+, CD335+, CD10+, BDCA-2+ cells (CD66b, CD335, CD10, BDCA-2 versus CD123). Lin−BDCA−CD123−/lo and Lin−BDCA−CD123hi populations are further subgated on the basis of the expression of CD34 versus CD117, CD135 versus CD116 and CD45RA versus CD115. In these gated populations, GMDPs are CD123−/lo CD34+CD117+CD135+CD116−CD115−CD45RA+; MDPs are CD123−/loCD34+CD117+CD135+ CD116−CD115+CD45RA+; CDPs are CD123hiCD34+ CD117+CD135+CD116+CD115−CD45RA+; and pre-cDCs are CD123−/loCD34−CD117+ CD135+CD116+CD115−CD45RA+SSC-Alo. FSC, forward scatter; SSC, side scatter.
Figure 5.
Gating strategy for culture output after hematopoietic differentiation on MS-5 stromal cells of human DC progenitors. (a) Workflow for isolation of DC progenitors. (b) CD34+ HSPCs from BM were isolated as in Figure 3c, and then cultured in MS5+FSG for 7 d. Culture output was assessed by flow cytometry using antibody panel #4, as indicated in Table 2. Cells developing in culture are identified by first gating on live cells (FSC-A versus SSC-A), and then on singlets (SSC-A versus SSC-W) and live cells (Live/Dead blue versus SSC-A). CD45+Lin− cells are obtained by successively gating on CD45+ cells (CD45 versus SSC-A) and gating out CD3+ cells (CD3 versus SSC-A), CD19+ cells (CD19 versus SSC-A) and CD335+ cells (CD335 versus SSC-A). Flow cytometry plots show phenotype of output cells, including granulocytes (brown), BDCA-3hi cDCs (red), BDCA-1+ cDCs (blue), monocytes (orange) and BDCA-2+ pDCs (green).
Figure 6.
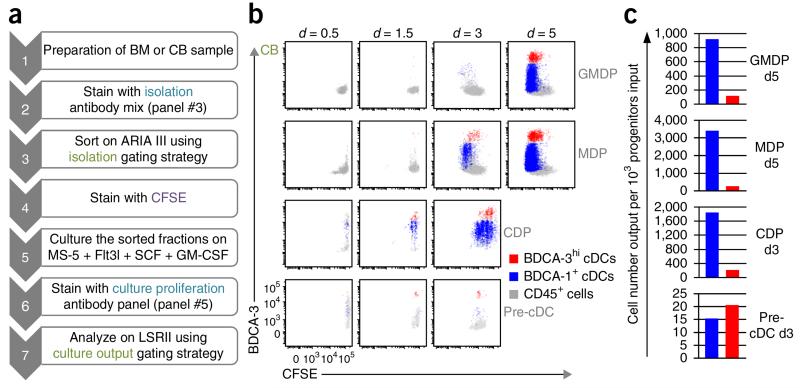
Proliferative capacity of human DC progenitors after hematopoietic differentiation on MS-5 stromal cells. (a) Workflow for culture proliferation of DC progenitors. (b) GMDPs, MDPs, CDPs and pre-cDCs were purified from CB, as shown in Figure 3c, labeled with CFSE, and cultured in MS5+FSG for 0.5, 1.5, 3 and 5 d; proliferation was assessed by flow cytometry using antibody panel #5, as indicated in Table 2. Flow cytometry plots show gated culture-derived cells, as defined in Figure 5b: CD45+ cells (gray), BDCA-1+ cDCs (blue) and BDCA-3hi cDCs (red). (c) Graphs showing the output of BDCA-1+ cDCs (blue) and BDCA-3hi cDCs (red) for 103 GMDPs (5 d), MDPs (5 d), CDPs (3 d) and pre-cDCs (3 d) cultured in MS5+FSG, as illustrated in Figure 6b.
Source tissue
The procedure can be performed on BM or CB samples. BM samples are typically obtained from elderly human donors; however, as age-related developmental changes may affect the composition of the progenitor compartment, CB cells can also be examined. As CB can be collected quite easily and cryopreserved, it is a good source of human progenitors. When these populations are purified and cultured in MS5+FSG, CB progenitors showed a differentiation potential similar to that exhibited by their BM counterparts14,15.
Identification and characterization of DC progenitors
As described, human DC progenitors can be identified by the lack of lineage and BDCA markers and by combining CD123, CD34, CD115, CD116, CD45RA, CD117 and CD135 markers (Table 2; panel #1). As shown in Figure 2b for BM and Figure 2c for CB, the following cells can be distinguished:
Cells with the phenotype Lin−BDCA−CD123−/loCD34+CD117+ CD135+CD116−CD115−CD45RA+ correspond to the GMDPs with the ability to generate granulocytes, monocytes and DCs.
Cells with the phenotype Lin−BDCA−CD123−/loCD34+CD117+ CD135+CD116− CD115+CD45RA+ represent the MDPs, which have the potential to give rise to monocytes and DCs but not to granulocytes.
Cells with the phenotype Lin−BDCA−CD123hiCD34+CD117+C D135+CD116+CD115−CD45RA+ correspond to the CDPs with the capacity to give rise to all DCs but not to monocytes and granulocytes.
Pre-cDCs have the phenotype Lin−BDCA−CD123−/loCD34− CD117+CD135+CD116+CD115−CD45RA+ and are restricted to the two subsets of cDCs.
Of note, we do not include isotype controls in our immunostaining, as the use of fluorescent isotype controls is not completely accurate, and because we have ascertained that the monoclonal antibodies are specific by carefully titrating them and using fluorescence minus one (FMO) controls for gating.
In nonhematopoietic tissues, we recommend using CD45 to visualize hematopoietic progenitors, as it may be challenging to identify these rare cells among other cells such as autofluorescent macrophages. As shown in Figure 2d, most, if not all, DC progenitors express CD45. Therefore, one channel in antibody panel #1 remains open to the potential use of CD45.
As DC progenitors are rare cells in both BM and CB, their identification is a matter of carefully defining the gates separating positive and negative cells. As shown in Figure 2b,c, the first step in the identification of human DC progenitors consists of excluding dead cells and debris by scatter gating. Live cells are then gated for singlets (side scatter(SSC)-W versus SSC-A; W = width, A = area), and dying cells are excluded on the basis of the uptake of the Live/Dead blue stain. The next step consists in the sequential exclusion of all lineage (CD3, CD19, CD14, CD66b, CD335 and CD10)- and BDCA (BDCA-1, BDCA-2 and BDCA-3)-expressing cells. To have a better resolution in our gates, we avoid as much as possible the use of a ‘dump’ channel containing all antibodies marking all excluded populations. Nonetheless, if the use of a ‘dump’ channel is considered to accommodate an additional antibody of interest (as done in Table 2, panel #3), one should carefully titrate each antibody separately before combining them to avoid any compensation issues and difficulties in discriminating between negative cells and cells with low expression. The use of CD123 allows clear separation of cells that express a high level of CD123 (CD123hi) from cells that express no or a low level of CD123 (CD123lo cells). CDPs are found within the CD123hi cells by sequentially gating on CD135+, CD116+, CD45RA+ and CD115− cells. The CD123lo gate is further analyzed by gating on CD34+ CD117+ (CD34+) cells or CD34− CD117+ (CD34−) cells. Pre-cDCs are found in the CD34− gate. Indeed, we first gate on cells that express CD135, CD116 and CD45RA but do not express CD115, and then exclude SSC-A-high cells, as these cells contain some monocyte precursors15. In contrast, the CD34+ population contains both GMDPs and MDPs. These progenitors are all CD135+, CD116− and CD45RA+; expression of CD115 allows the separation of GMDPs (CD115− cells) from MDPs (CD115+ cells). Even though the CD115 staining is very faint and it is not possible to get a clear-cut separation of positive and negative cells, the use of FMO control allows for proper gating on CD115+ cells (Fig. 2e). Moreover, if we consider that lineage commitment occurs when the capacity to give rise to one cell type, e.g., monocyte and DC, is increased and the capacity to produce another cell type, e.g., granulocyte, is lost, dim CD115 expression is sufficient to separate cells with GMDP potential from cells with MDP potential.
To characterize DC progenitors, we use a related combination of antibodies (Table 2, panel #2). Panel #2 leaves a channel open to accommodate an additional antibody of interest. We prefer the phycoerythrin (PE) channel for this purpose, as PE is bright, and most if not all antibodies are available in this color. Because CD135 staining is crucial in order to identify DC progenitors and because only CD135 PE works well in our hands, we decided to open the PE-Cy7 channel. PE-Cy7 is a tandem dye that can degrade in the presence of light and heat, and it will then emit in the parent dye channel (PE). However, by avoiding the exposure of samples to light and heat, this problem can be largely avoided. The gating procedure is the same as that previously described (Fig. 2b,c), and the expression of CD11c, HLA-DR, CD38 and CD33 on DC progenitors is shown (Fig. 3b). HLA-DR, whose expression is neither specific nor identical within the DC subsets, is also expressed by DC progenitors. CD11c, which is weakly expressed on cDCs and completely lacking on pDCs, is upregulated during DC development. CD38, which was initially used to identify multipotential progenitors16, as well as DC lineage–committed progenitors14, is also expressed by pre-cDCs. Because CD38 is expressed by all hematopoietic progenitors, we decided not to use it in our antibody panels and focus more on hematopoietin receptors that allow further separation of these cells. CD33, which is highly specific to the hematopoietic compartment, is expressed by all DC progenitors.
Isolation of progenitors
We developed our flow cytometry strategy to identify DC progenitors on a 16-color flow cytometer (BD LSR II with blue, yellow/green, red, violet and UV lasers; see configuration in Table 3). To isolate DC progenitors, we adapted our panel to maximize the capacities of an 11-color cell sorter (BD Aria III with blue, yellow/green, red and violet lasers; see configuration in Table 4). This was done in part by restricting the number of channels used for lineage markers and by omitting the Live/Dead blue stain to exclude dead cells, relying only on exclusion by scatter gating (Table 2, panel #3). Moreover, the gating hierarchy while sorting is limited to a maximum of an eight-gate sort depth. This limit is intrinsic to the current generation of BD Aria III sorter hardware and firmware. To limit our gating strategy to eight steps (Fig. 4b), we also removed the step to gate out the singlets (SSC-W versus SSC-A). The gating strategy to identify and sort all DC-defined progenitors simultaneously is shown in Figure 4c. The sorted cells are suitable for various downstream applications such as cell culture or gene expression profiling.
Composition and proliferative capacity of in vitro–differentiated cells
Hematopoietic differentiation can be induced in vitro by coculture of hematopoietic progenitors on mouse BM stromal cells plus cytokines. To assess the differentiation potential of different hematopoietic progenitors and their multilineage or restricted potential, we developed an additional antibody panel (Table 2, panel #4). BM CD34+ HSPCs cultured in MS5+FSG for 7 d can produce T cells (CD3+), B cells (CD20+), NK cells (CD335+), granulocytes (CD66b+), monocytes (CD14+) and all three major subsets of DCs, that is, pDCs and both subsets of cDCs (Fig. 5b). Human CD45 antibody must be included in this panel, as it discriminates between human cells and mouse stromal cells, which are not entirely gated out by scatter analysis. Of note, BDCA markers are often used to classify human DCs. However, they are not strictly restricted to DCs; we therefore used them in combination with Clec9A for BDCA-3hi cDCs, CD14 for BDCA-1+ cDCs and CD123 for BDCA-2+ pDCs.
To examine the proliferative potential of DC progenitors, we purified them from CB and stained them with carboxy-fluorescein diacetate succinimidyl ester (CFSE) before culturing in MS5+FSG. By using a 14-color antibody panel (Table 2, panel #5) that includes CFSE, we were able to track the cells that were expanding in culture by CFSE dilution. As shown in Figure 6b,c, all progenitors are expanding in culture, even though DC lineage–committed progenitors—GMDP, MDP and CDP cells—have a greater proliferative capacity than DC immediate precursors, i.e., pre-cDCs. Moreover, BDCA-1+ and BDCA-3hi cDCs differentiate as early as 12 h from CDP and pre-cDC progenitors.
Applications of the protocol
We believe that the protocol described here has multiple applications and a wide interdisciplinary scope including (i) further definition of human DC hematopoiesis in health, disease and vaccine settings, as well as (ii) evaluating DC progenitors’ applications in therapeutic approaches.
Our protocol can be used as an initial tool to analyze normal or abnormal immunophenotypic changes during DC development in humans. For instance, it can facilitate the study of diseases involving aberrant DC hematopoiesis such as chronic myelogenous leukemia (CML)21 or a newly identified form of primary immunodeficiency characterized by recurrent infections and disseminated bacille Calmette-Guérin (BCG) infection after vaccination in patients lacking GATA2 (ref. 22) and interferon regulatory factor 8 (IRF8)23. Because of their capacity to elicit and regulate immune responses, DCs are essential to any effort for vaccine development. Therefore, our protocol provides important tools to design and monitor and potentially improve vaccines that rely on DCs.
Finally, the methods described above can support the exploration of new cellular therapeutic approaches based on DC progenitors including stem cell therapies focused on embryonic stem cells and induced pluripotent stem cells. The composition and the properties of the cells that are produced after in vitro differentiation of progenitors will differ considerably depending on the culture condition, and therefore this system can be used to evaluate DC progenitors to identify novel regulators of DC hematopoiesis or to test the toxicity of new drug candidates. Of note, therapeutic approaches based on human DC progenitors may require the development of a nonxenogeneic culture system; therefore, it would be good to explore the feasibility of expanding DC progenitors in an entirely human coculture system using either umbilical vein endothelial cells for CB progenitors or BM stromal cells for BM progenitors.
MATERIALS
REAGENTS
Cell samples
Human samples can be obtained from national blood centers for CB and from local hospitals for BM ! CAUTION Universal precautions must be taken while manipulating human specimens, and all experiments must be carried out in at least class II biological safety cabinets and using appropriate protection equipment.! CAUTION Parental informed consent (CB) or patient informed consent (BM) must be obtained before collection, and protocols must conform to your Institutional Review Board (IRB)’s guidelines. IRB-approved protocols usually require that the samples be anonymous and fully deidentified at the site of collection by hospital personnel.
Cell culture
Dulbecco’s PBS 1× (DPBS; Life Technologies, cat. no. 14190)
RPMI-1640 (Life Technologies, cat. no. 21870-076)
α-MEM 1× (Life Technologies, cat. no. 12561-056)
-
FBS, heat inactivated (Life Technologies, cat. no. 16140-071)
▲ CRITICAL We recommend testing several lots of serum for culture optimization and then using the same lot for an entire series of experiments.
In our case, we always test three or four lots for hematopoietic differentiation on MS-5 stromal cells and select the one that supports the development of CD34+ HSPCs with an overall better yield for DCs.
Antibiotic-antimycotic solution (Cellgro, cat. no. 30-004-CI)
Collagenase type IV (Sigma-Aldrich, cat. no. C-5138)
Ficoll-Paque plus (GE Healthcare, cat. no. 17-1440-03)
Trypsin-EDTA, 0.05% (Life Technologies, cat. no. 25300-054)
EDTA, 0.5 M (Life Technologies, cat. no. 15575)
ACK lysing buffer (Life Technologies, cat. no. A10492)
Mitomycin C (Sigma-Aldrich, cat. no. M4287) ! CAUTION Avoid contact and inhalation. Wear gloves, safety glasses and a lab coat.
DMSO (Sigma-Aldrich, cat. no. 154938)
Trypan blue stain 0.4% (wt/vol) (Life Technologies, cat. no. 15250-061)
Carboxy-fluorescein diacetate succinimidyl ester (CFSE; Life Technologies, cat. no. C1165)
Recombinant human SCF ligand (SCF; R&D systems, cat. no. 255-SC-050/CF)
Human FLT3 ligand (FLT3L; Celldex Therapeutics, special order)
-
Leukine (sargramostim) as a human GM-CSF (Genzyme)
▲ CRITICAL Leukine is a prescription drug that must to be ordered through a pharmacy.
Human serum albumin (HSA; Sigma-Aldrich, cat. no. A9511)
MS-5 mouse bone marrow stromal cell line (Creative Bioarray, cat. no. CSC-C2763) ! CAUTION The cell lines used in your research should be regularly checked to ensure that they are authentic and that they are not infected with mycoplasma.
Benzonase nuclease, 10 kU, 25 U/μl (Novagen, cat. no. 70664-3)
Heparin, cell culture grade (Sigma-Aldrich, cat. no. H3149)
Bleach ! CAUTION Bleach is an irritant. Avoid contact with eyes, skin and clothing.
Plasticware
50-ml conical tubes (BD Falcon, cat. no. 352098)
15-ml conical tubes (BD Falcon, cat. no. 352097)
5-ml polypropylene round-bottom tube (BD Falcon, cat. no. 352063)
5-ml polystyrene round-bottom tube (BD Falcon, cat. no. 352052)
5-ml polystyrene round-bottom tube with cell-strainer cap (BD Falcon, cat. no. 352235)
25-ml serological pipette (Costar, cat. no. 4489)
10-ml serological pipette (Costar, cat. no. 4488)
5-ml serological pipette (Costar, cat. no. 4487)
Transfer pipette (Sarstedt, cat. no. 86.1171.001)
Microcentrifuge tubes (1.5 ml; Eppendorf, cat. no. 022363204)
Cryogenic screw-cap micro tubes (2-ml; Sarstedt, cat. no. 72.694.006)
Micro tubes with screw caps, conical bottom (1.5-ml; RPI Corp, cat. no. 144500)
Titertube micro test tubes (Bio-Rad, cat. no. 223-9391)
96-well micro-well plates V-bottom without lid (Nunc, cat. no. 249570)
96-well micro-well plates lid (Nunc, cat. no. 263339)
96-well tissue culture plates flat-bottom (BD Falcon, cat. no. 357072)
10-cm tissue culture dishes (BD Falcon, cat. no. 353003)
15-cm tissue culture dishes (BD Falcon, cat. no. 353025)
150-cm2 tissue culture flasks straight neck with vented cap (BD Falcon, cat. no. 355001)
10-ml syringe (BD Falcon, cat. no. 309604)
Syringe filter with a 0.2-μm membrane (Pall, cat. no. 4612)
Cell strainer, 100 μm (BD Falcon, cat. no. 352360)
Cell strainer, 70 μm (BD Falcon, cat. no. 352350)
Precision specimen container, 4 ounce, positive seal indicator (Covidien, cat. no. 2200SA)
Flow cytometry reagents
Antibodies (Table 1) ▲ CRITICAL Each antibody must be titrated before use (see Reagent Setup) ! CAUTION All antibodies must be kept at 4 °C and protected from exposure to direct light.
Live/Dead fixable blue dead cell stain kit for UV excitation (Life Technologies, cat. no. L23105) ! CAUTION Keep it at −20 °C and protect it from exposure to direct light.
Formaldehyde solution 37% (wt/vol) H2O (Sigma-Aldrich, cat. no. 252549) ! CAUTION Formaldehyde is toxic by inhalation or if swallowed; it is irritating to the skin, eyes and respiratory system, and it may be carcinogenic. Formaldehyde should be used with appropriate safety measures, such as protective gloves, glasses, clothing and sufficient ventilation. All waste should be handled according to hazardous waste regulations.
BD CompBeads set anti-mouse Igκ (BD Biosciences, cat. no. 552843)
BD CompBeads set anti-rat Igκ (BD Biosciences, cat. no. 552844)
Amine-reactive compensation bead kit (Life Technologies, cat. no. A10346)
EQUIPMENT
Biological safety level 2 (BSL-2) tissue culture facilities
BSL-2 sorting facilities
Class II biological safety cabinet (The Baker Company, cat. no. SG403)
Cell incubator (37 °C, 5% CO2; Nuaire, cat. no. NU-5500)
Tabletop centrifuge with rotors and adapters for 50- and 15-ml conical tubes and 96-well plates (Sorvall, cat. no. 75-004-367)
Microcentrifuge (Eppendorf, cat. no. 5424)
Inverted microscope (Nikon, cat. no. Eclipse TS100)
Hemocytometer (Fisher Scientific, cat. no. 02-671-5) or other cell counter
Cryo 1 °C freezing container (Nalgene, cat. no. 5100-0001)
LN2 storage systems (Thermo Scientific, cat. no. 7404)
BD LSR II flow cytometer running DIVA 8.0, or equivalent (BD Biosciences, special order). Our LSR II is equipped with four lasers with a configuration allowing 16-color, 18-parameter flow cytometry (Table 3)
BD Aria III cell sorter running DIVA 8.0, or equivalent (BD Biosciences, special order). Our Aria III is equipped with three lasers with a configuration allowing 11-color, 13-parameter flow cytometry (Table 4)
BD FACSDiva V8.0.1 software (BD Biosciences) for data acquisition
FlowJo software V9.8.2 (TreeStar) for data analysis
REAGENT SETUP
Heparin
Dissolve heparin in PBS at a final concentration of 1,000 U/ml. Filter this solution with a 0.2-μm syringe filter and divide the solution into aliquots in a 5-ml polypropylene round-bottom tube. Store the heparin at 4 °C for up to 2 years.
Collagenase solution
Dissolve 100 mg of collagenase type IV in 50 ml of RPMI-1640 to obtain a 2 mg/ml collagenase stock solution. Filter this solution with a 0.2-μm syringe filter. Store the solution in 10-ml aliquots at −20 °C for up to 1 year.
Freezing medium
Prepare freezing medium by adding 10% (vol/vol) DMSO in FBS on the day of use.
Thawing medium
Prepare thawing medium by adding 10% (vol/vol) FBS in RPMI-1640, and store it at 4 °C for up to 4 weeks.
CFSE stock
CFSE is resuspended in DMSO at a concentration of 5 mM. Store the stocks in aliquots of 50 μl in Eppendorf tubes at −80 °C for up to 1 year. Each batch of CFSE should be titrated before use in the procedure (Box 1) on cells to determine the optimal concentration for labeling (0.5 μM final was used in the example results shown here).
Box 1. CFSE staining ● TIMING 30 min.
To assess the ability of cells to proliferate in culture (Steps 36–38), we use a cell proliferation assay. Cells are labeled with the fluorescent dye CFSE. Those cells that proliferate show a reduction in CFSE fluorescence intensity, which is measured directly by flow cytometry.
Wash the cells twice in PBS and resuspend them in PBS at a 2 × 107 cells per ml concentration.
Prepare a 1 μM working solution of CFSE from the stock (5 mM) by diluting the stock in PBS. Label the cells by adding one volume of working solution of CFSE (1 μM) to one volume of cells. The final concentration of CFSE is then 0.5 μM.
Incubate the cells in the dark with gentle mixing at room temperature for 8 min.
Quench the reaction by adding an equal volume of FBS for 2 min.
Wash the cells twice with PBS and resuspend them in MS-5 growth medium.
To assess the proliferation of bulk CD34+ cells or purified progenitors (see Step 36B for isolation of progenitors using panel #3) during hematopoietic differentiation on MS-5 stromal cells, proceed directly to Step 33.
MS-5 growth medium
Prepare the growth medium by adding 10% (vol/vol) FBS and 1% (vol/vol) penicillin-streptomycin in α-MEM.
▲ CRITICAL Prepare the medium in advance and store it at 4 °C for up to 2–3 weeks.
Mitomycin C solution
Dissolve mitomycin C in sterile H2O to obtain a 0.5 mg/ml stock solution. Store the solution in 500-μl aliquots at −20 °C for up to 1 year.
Cytokine buffer
Mix 0.1% (vol/vol) HSA in PBS. Sterilize the solution by filtration using a 0.2-μm syringe filter, and store it for up to 6 months at 4 °C.
Reconstitution of cytokines
Reconstitute human cytokines in cytokine buffer at a stock concentration of 20 μg/ml for GM-CSF and SCF ligand and 100 μg/ml for FLT3L. Store the cytokines in 50-μl aliquots at −80 °C for up to 2 years. ▲ CRITICAL Vials of lyophilized cytokines should be centrifuged in a microcentrifuge at maximum speed for 1 min at room temperature (20–25 °C) before opening vials.
Formaldehyde working solution
Mix 2% (vol/vol) formaldehyde in PBS. Store the solution at 4 °C for up to 1 month.
Staining buffer
Mix 1% (vol/vol) FBS in PBS. ▲ CRITICAL Staining buffer can be prepared in advance and stored at 4 °C for up to 1 month.
Sorting buffer
Mix 1% (vol/vol) FBS and 0.5 M EDTA (2 mM final) in PBS. ▲ CRITICAL Sorting buffer can be prepared in advance and stored at 4 °C for up to 1 month.
Live/Dead blue viability dye
Thaw Live/Dead blue powder by leaving it for 10 min at room temperature. Add 50 μl of DMSO and vortex it thoroughly. Store the solution in 1-μl aliquots in Eppendorf tubes at −20 °C for up to 1 year. Each batch of Live/Dead blue should be titrated before use in the procedure on dying cells, for example, cells kept in culture at a very dense concentration such as 20 × 106 cells per ml, to determine the optimal dilution for labeling (1/1,000 final in the example results shown here). When you are ready to use Live/Dead blue, thaw and keep on ice a 1-μl aliquot into which you add 99 μl of PBS (1/100 Live/Dead blue working solution).
Amine-reactive Live/Dead blue CompBeads
Vortex the beads thoroughly before use. Resuspend one drop of positive beads and one drop of negative beads in 500 μl of PBS. Add 20 μl of Live/Dead blue working solution to 180 μl of beads solution in a 5-ml polypropylene round-bottom tube (final dilution of Live/Dead blue is 1/1,000). Incubate the mixture for 30 min at 4 °C in the dark. Wash the beads twice with PBS. Resuspend the beads in 150 μl of formaldehyde working solution and transfer them to a titer tube. Prepare the beads on the day of use. ▲ CRITICAL It is crucial to use PBS alone and not staining buffer that contains serum, as proteins from the serum would prevent the binding of Live/Dead blue to the compensation beads.
Antibody staining solution
The amounts of antibody needed per staining are determined by titration experiments done before performing the procedure. The optimal concentration of an antibody is determined by a titration experiment in which typically 2 × 106 cells in a 20-μl volume are stained with 1×, 2×, 4× and 8× the amount of antibody. The optimal concentration for a specific antibody is the amount giving the best fluorochrome signal and the least fluorochrome spreading. To prepare the antibody staining solution, mix the titrated amount of antibody for all antibodies and make it up to 20 μl with staining buffer (Table 2). The staining volume is 40 μl total (20 μl of cell suspension + 20 μl of antibody staining solution). The staining volume should be scaled up according to the number of cells. Prepare this solution on the day of use. ▲ CRITICAL Spin the antibody staining solution in a microcentrifuge at maximum speed for 1 min at room temperature to remove antibody aggregates. Use the supernatant only to stain cells. ▲ CRITICAL Always prepare the antibody staining solution in excess (10% more than needed for the experiment).
BD CompBeads
Vortex the beads thoroughly before use. Prepare a positive bead solution and a negative bead solution (typically one drop of beads is added to 500 μl of PBS for the positive solution). For the negative FACS control, add 100 μl of negative bead solution in one well of a 96-well V-bottom plate. For the positive FACS controls, add 100 μl of positive bead solution per well of a 96-well V-bottom plate for each control. Add the titrated amount of antibody. Incubate the 96-well plate for 30 min at 4 °C in the dark. Wash the beads twice with PBS. Resuspend each control in 150 μl of formaldehyde working solution. Transfer the controls to titer tubes. Prepare the beads on the day of use. ▲ CRITICAL Carefully check that the monoclonal antibody chosen can be recognized by the beads (for example, use anti-mouse Igκ CompBeads for mouse monoclonal antibodies).
PROCEDURE
Donor CB and BM collection and initial processing ● TIMING 10–45 min
! CAUTION All human blood, body fluids and tissues must be treated as if infectious and universal precautions must be taken by lab workers while manipulating these specimens.
! CAUTION All human biological waste to be discarded should be treated with bleach or it should be autoclaved, and then it can be discarded according to your institution disposal procedures.
▲ CRITICAL All steps should be carried out in a class II biological safety cabinet.
-
1|
Obtain human samples from CB, as described in option A, or obtain and prepare a single-cell suspension of BM, as described in option B.
(A) CB sample ● TIMING ~10 min
-
(i)
Obtain a fresh cord blood unit (CBU; typically 80–140 ml) provided in citrate phosphate dextrose adenine (CPDA-1) preservative solution.
▲ CRITICAL STEP The CBU should be kept at room temperature until processing. For optimum results, processing should take place in <12 h, and the samples that are older than 24 h should not be used.
(B) BM sample ● TIMING ~45 min
-
(i)
Obtain biopsies from surgical procedures such as routine hip arthroplasty. The biopsy should be kept on ice immediately after surgical removal in a container for specimen collection, and it should be preserved in 1–2 ml of heparin (1,000 units per ml).
-
(ii)
Weigh the specimen for reference.
▲ CRITICAL STEP Samples can be stored at 4 °C for a few hours before starting the isolation procedure.
▲ CRITICAL STEP This protocol has been optimized for solid biopsies and not aspirates—i.e., a sample of the liquid portion of BM.
▲ CRITICAL STEP This protocol is optimized for the processing of specimen weighing 20–40 g. For smaller or larger specimens, adjust the volumes accordingly.
-
(iii)
Wash the BM biopsy in PBS and transfer it into a 50-ml tube with 10–20 ml of collagenase solution. Incubate the mixture for 30 min at 37 °C under continuous agitation.
! CAUTION Collagenase digestion of tissues to create single-cell suspensions may result in the destruction of certain cell surface markers. However, we did not observe the destruction of any of the surface markers that we tested.
▲ CRITICAL STEP Add 10 ml of collagenase solution to every 20 g of tissue. Collagenase is thawed and kept on ice.
▲ CRITICAL STEP This short digestion allows the disruption of the bony matrix, as well as the release of the fat constituting the marrow.
-
(iv)
Filter the BM cell suspension through a fine mesh nylon screen.
Isolation of CB and BM MNCs via density gradient ● TIMING 1.5 h
-
2|
Prepare CB (option A) or BM (option B) density gradients.
(A) CB gradient
-
(i)
Dilute the entire CBU 1:1 with sterile PBS in an upright appropriately sized sterile tissue culture flask.
(ii) Carefully and slowly layer 35 ml of diluted CB onto 15 ml of Ficoll-Paque in a 50-ml conical tube. Repeat the process in additional tubes for the remaining diluted CBU. If the last amount does not total 35 ml, make up the volume to 35 ml with PBS.
(B) BM gradient
-
(i)
Carefully and slowly layer 35 ml of BM cell suspension onto 15 ml of Ficoll-Paque in a 50-ml conical tube.
If the amount does not total 35 ml, make up the volume to 35 ml with PBS.
▲ CRITICAL STEP The more diluted the sample, the better the purity of the MNCs.
▲ CRITICAL STEP To avoid mixing Ficoll-Paque and the cell suspension, tilt the 50-ml conical tube at a 45 °C angle and slowly let the blood run down on the side of the tube onto the Ficoll layer.
-
3|
Centrifuge the mixture at 850g for 30 min at 20 °C in a swinging-bucket rotor with no brake.
-
4|
Aspirate the upper layer (yellow layer corresponding to the plasma, in the case of CB) leaving the mononuclear cell layer (white layer at the interphase between the Ficoll and the plasma) undisturbed.
? TROUBLESHOOTING
-
5|
Carefully transfer each mononuclear cell layer using a sterile transfer pipette to a new 50-ml conical tube.
-
6|
Fill the 50-ml conical tubes with RPMI-1640, mix well, and centrifuge the tubes at 480g for 5 min at 20 °C with brake.
-
7|
Carefully remove the supernatant completely. Pool the pellet of all 50-ml conical tubes. Fill the 50-ml conical tube with RPMI-1640, mix it well and centrifuge it at 480g for 5 min at 20 °C with brake.
▲ CRITICAL STEP CB samples sometimes show substantial contamination of the interphase layer with red blood cells. In that case, resuspend the cell pellet in 5 ml of ACK lysing buffer. Allow it to stand for 5 min at 4 °C. Fill the 50-ml conical tube with RPMI-1640, mix it well and centrifuge it at 480g for 5 min at 20 °C with brake. Remove the supernatant; resuspend the cells in 50 ml of RPMI-1640 and centrifuge them once more at 480g for 5 min at 20 °C with brake.
-
8|
Carefully remove the supernatant completely. Resuspend the cells in 50 ml of staining buffer, count the cells using a hemocytometer or other cell counter, and determine the viability by trypan blue stain exclusion. If you are not pausing, proceed directly to Step 35 (staining panel #1, 2 or 3) or Step 33 (staining panel #4 or 5).
■ PAUSE POINT Keep the cells in staining buffer at 4 °C for 1–2 h until use, or freeze them for long-term storage, as described in Steps 9–12.
Freezing of CB and BM MNCs ● TIMING 20 min
-
9|
Centrifuge the cells at 480g for 5 min at 20 °C.
-
10|
Resuspend the cells at a density of 10 × 106 cells per ml in freezing medium.
-
11|
Transfer 1 ml of cells in freezing medium per sterile cryogenic micro tube.
▲ CRITICAL STEP Label each tube clearly (date, sample name, cell concentration).
▲ CRITICAL STEP The optimal cell concentration for freezing is 10 × 106 cells per ml. However, you can go up to 50 × 106 cells per ml in case of large specimens.
-
12|
Put the cryogenic vial in a freezing container, and store it at −80 °C for 12–24 h before transferring it to the liquid nitrogen tank for long-term storage.
■ PAUSE POINT Cells can be stored in liquid nitrogen for years.
Thawing CB and BM MNCs ● TIMING 30 min
-
13|
Add 10 ml of cold thawing medium in a 15-ml conical tube to thaw one vial or add 40 ml of cold thawing medium in a 50-ml conical tube to thaw up to ten vials.
▲ CRITICAL STEP To obtain the best possible survival, thawing of cells must be performed as quickly as possible, and the thawing medium should be cold to minimize heat shocks.
-
14|
Remove the vial from the liquid nitrogen tank, and hold it at room temperature until the sides are thawed but the center remains frozen.
▲ CRITICAL STEP Do not place the vial into a 37 °C water bath, as it is important not to dramatically heat-shock the cells.
-
15|
Transfer the contents of the vial (~1 ml) into a 15-ml conical tube containing cold thawing medium.
-
16|
Centrifuge the cells at 480g for 5 min at 20 °C.
-
17|
Resuspend the cells in staining buffer for extemporaneous use.
Culture of mouse MS-5 stromal cells ● TIMING 8 d
▲ CRITICAL Mouse MS-5 stromal cells are only required for staining panel #4 or 5.
-
18|
Culture MS-5 stromal cells in 10-cm tissue culture dishes with 10 ml of MS-5 growth medium. Place the cells in a CO2 cell incubator.
▲ CRITICAL STEP MS-5 stromal cells grow in monolayers. Examine the tissue culture dish under the microscope; the cells are ready to be split when they are 95% confluent.
-
19|
Aspirate the MS-5 growth medium and wash the cells twice with 10 ml of PBS.
-
20|
Add 2 ml of trypsin-EDTA 0.05% (wt/vol) solution and incubate the cells for 2 min at 37 °C in a CO2 cell incubator.
▲ CRITICAL STEP Inadequate washing of cells with PBS or using an old trypsin solution may result in partial detachment of cells from the dish.
-
21|
Add 10 ml of MS-5 growth medium and collect the cells by pipetting.
-
22|
Transfer the cell suspension in a 15-ml conical tube and centrifuge it at 480g for 5 min at 20 °C.
-
23|
Aspirate the supernatant and resuspend the cells in 10 ml of MS-5 growth medium.
-
24|
Add 0.5 ml of cell suspension to 9.5 ml of MS-5 growth medium, and plate the cells onto a 10-cm tissue culture dish. Place the cells in a CO2 cell incubator.
-
25|
When the culture is 95% confluent, if required, split the dish for maintenance (by repeating Steps 19–24). Confluence should be reached after 3–4 d of culture.
-
26|
To expand the cells for coculture experiment, prepare an extra tissue culture dish by adding 1 ml of cell suspension to 19 ml of MS-5 growth medium into a 15-cm Petri dish.
▲ CRITICAL STEP Do not use cells that have a high passage number or that are growing slowly. The doubling time for healthy MS-5 is ~24 h.
■ PAUSE POINT It is possible to pause the protocol at this stage by freezing and storing MS-5 stromal cells in liquid nitrogen tank for future use. Cells can be frozen and thawed as described in Steps 9–17 for MNCs.
Hematopoietic differentiation on MS-5 stromal cells ● TIMING 8 d
-
27|
The day before the coculture experiment (day 0), take the extra tissue culture dish of MS-5 stromal cells that was prepared (Step 26), and proceed to mitomycin C treatment. Aspirate the medium without disturbing the MS-5 stromal cell layer. Add 20 ml of fresh MS-5 growth medium containing 10 μg/ml mitomycin C. Incubate the dish for 3 h in a CO2 cell incubator.
-
28|
Trypsinize the MS-5 stromal cell layer, as described in Steps 19–23.
-
29|
Count the MS-5 cells using a hemocytometer or other cell counters and determine viability by trypan blue stain exclusion.
-
30|
Centrifuge the cells at 480g for 5 min at 20 °C; aspirate the supernatant.
-
31
Resuspend the pellet at 2.5 × 105 per ml in MS-5 growth medium.
-
32|
Seed 2.5 × 104 cells (100 μl) per well of a 96-well flat-bottom tissue culture plate. Incubate the cells overnight in a CO2 cell incubator.
-
33|
The following day (day 1), resuspend a maximum of 104 bulk CD34+ cells or purified progenitors (see Step 36B for isolation of progenitors using panel #3) in 100 μl of MS-5 growth medium containing 40 ng/ml GM-CSF and SCF ligand and 200 ng/ml FLT3L.
▲ CRITICAL STEP If progenitors need to be stained with CFSE (panel #4) before coculture with MS-5 stromal cells, proceed as described in Box 1.
-
34|
Seed the progenitor cells on top of the MS-5 stromal cells prepared in Step 32. Incubate the cells for 7 d in a CO2 cell incubator.
Cell surface staining ● TIMING 1–2 h
-
35|
Take all or an aliquot of the cell suspension from Step 8. Use at least 2–4 × 106 cells for simple phenotyping (panel #1) and characterization (panel #2) staining. If you are sorting the cells (panel #3), start with a sufficient number of cells to obtain a reasonable number of progenitors after sorting. We usually sort 100–200 × 106 CB or BM MNCs to achieve sufficient progenitors. Use the whole MS-5 culture from Step 34 for culture output (panel #4) and culture proliferation (panel #5) staining.
▲ CRITICAL STEP We recommend working with fresh samples. If you are working with frozen samples, thawing and staining should preferably be performed on the same day. If you are using frozen cells, count the cells (from Step 17) after thawing using the hemocytometer or other cell counter and determine the viability by Trypan Blue Stain exclusion.
? TROUBLESHOOTING
-
36|
If you are performing cell surface staining for panel #1 and 2, follow option A; for panel #3, follow option B; and for panel #4 and panel #5, follow option C.
! CAUTION This and all subsequent steps should avoid exposure to direct light to preserve fluorochromes.
▲ CRITICAL STEP Do not forget to prepare the compensation tubes as well (see Reagent Setup).
(A) Staining for panel #1 and #2 ● TIMING 1 h 30 min
-
(i)
Resuspend the cells (from Step 8) in PBS at a density of 100 × 106 cells per ml.
-
(ii)
Put 2 × 106 cells (20 μl) in each well of a 96-well V-bottom plate.
-
(iii)
Add 2 μl of 1/100 Live/Dead blue working solution to each well.
-
(iv)
Incubate the cells at 4 °C in the dark for 20 min.
-
(v)
Wash the cells by adding 150 μl of staining buffer per well.
-
(vi)
Centrifuge the cells at 480g for 5 min at 20 °C; flick the plate to decant the supernatant.
-
(vii)
Add 20 μl of the antibody staining solution per well.
-
(viii)
Incubate the cells at 4 °C in the dark for 30 min.
-
(ix)
Wash the cells by adding 150 μl of staining buffer per well.
-
(x)
Centrifuge the cells at 480g for 5 min at 20 °C; flick the plate to decant the supernatant.
-
(xi)
Resuspend the cells in 150 μl of formaldehyde working solution per well. Transfer the samples to titer tubes.
-
(xii)
Keep the samples in the dark at 4 °C, and acquire samples on a flow cytometer.
■ PAUSE POINT Fixed cells can be kept in the dark at 4 °C for up to 48 h before analyzing by flow cytometry.
(B) Staining for panel #3 ● TIMING 1 h
-
(i)
Resuspend the cells (from Step 8) in staining buffer at a density of 400 × 106 cells per ml in a 50-ml conical tube.
-
(ii)
Add 20 μl per 8 × 106 cells of the antibody staining solution.
-
(iii)
Incubate the cells at 4 °C in the dark for 30 min.
-
(iv)
Wash the cells by adding 40 ml of sorting buffer.
-
(v)
Centrifuge the cells at 480g for 5 min at 20 °C. Aspirate the supernatant.
-
(vi)
Resuspend the cells in sorting buffer at a density of 30 × 106 cells per ml, and transfer them to a 5-ml polypropylene round-bottom tube.
▲ CRITICAL STEP We recommend filtering the cell sample before sorting using a 5-ml polystyrene round-bottom tube with a cell-strainer cap. Transfer the sample in a 5-ml polypropylene round-bottom tube for sorting.
-
(vii)
Keep the sample in the dark on ice, and then sort the sample on a cell sorter in a timely manner (within 1 h).
(C) Staining for panel #4 and #5 ● TIMING 1 h 30 min
-
(i)
Collect all cells from MS-5 stromal coculture (from Step 34) by vigorous pipetting, and transfer them to a 96-well V-bottom plate.
-
(ii)
Centrifuge the cells at 480g for 5 min at 20 °C; flick the plate to decant the supernatant.
-
(iii)
Stain the cells by the standard surface staining method, as in Step 36A(iii–xii).
■ PAUSE POINT Fixed cells can be kept in the dark at 4 °C for up to 48 h before analyzing by flow cytometry.
Acquisition and cell sorting ● TIMING ~1–4 h
-
37|
Run the compensation controls (see Reagent Setup). Create the compensation matrix and apply it to tubes for both acquisition and sorting.
▲ CRITICAL STEP Compensation controls must consist of a negative and a positive population for each single fluorochrome. The negative and positive populations should have the same autofluorescence, which means that each positive bead should be compared with its matching negative bead.
▲ CRITICAL STEP When you first set up a panel, you must include a nonstained tube, as well as FMO control (FMOC) tubes. FMOCs sequentially contain all fluorochrome or tandem dyes minus one; they are used to define proper gates and to control the compensation process.
? TROUBLESHOOTING
-
38|
Run the samples (from Step 36A(xii) and Step 36C(iii)) for acquisition on the flow cytometer. Analyze data using FlowJo, as depicted in Figure 2 (panel #1), Figure 3 (panel #2), Figure 5 (panel #4) and Figure 6 (panel #5).
▲ CRITICAL STEP If you do not have FlowJo software, use FACSDiva software or an equivalent.
? TROUBLESHOOTING
-
39|
Proceed to simultaneous sorting of the four populations of interest—i.e., GMDPs, MDPs, CDPs and pre-cDCs—from MNCs obtained in Step 8 and stained in Step 36B(i-vi) using an 85-μm nozzle cell sorter. Collect the sorted cells in 1.5-ml conical-bottom tubes with screw caps, in which you add 300 μl of MS-5 growth medium and vortex the collection tubes to coat the surface. You can collect up to 2.5 × 105 cells per tube.
▲ CRITICAL STEP If you are sorting cells to analyze mRNA expression by gene array, cells should be recovered without interruption and should be kept at 4 °C at all times to avoid changes in gene expression.
? TROUBLESHOOTING
? TROUBLESHOOTING
Step 4
If the upper layer is contaminated with RBCs, the samples—that is, upper layer and interphase—should be mixed and layered over a second batch of Ficoll in a new 50-ml conical tube. Repeat this step if necessary.
Step 35
Post-thaw samples may clot. The cells are probably aggregating because of the release of DNA by cells dying during the freezing-thawing process. Include DNase such as benzonase (final concentration 25 U/ml) in thawing medium (Steps 13–17), as well as in staining buffer in Steps 36A(iii,iv), 36A(vii,viii) and 36B(ii,iii).
Step 37
If you have no staining of compensation beads, it may be because not all compensation controls have been prepared in Step 36. You must prepare a compensation control for each fluorochrome or conjugate if you are using tandem dyes.
It may also be that you are not using the proper beads; use anti-mouse Igκ for mouse monoclonal antibodies and anti-rat Igκ for rat monoclonal antibodies.
Step 38
If you have clogging of the sample line while acquiring samples, the cells are probably aggregating because of poor viability. We recommend resuspending the cells in sorting buffer that contains EDTA and filtering the cells using 5-ml polystyrene round-bottom tubes with cell-strainer caps.
Step 39
If you have clogging of the sample line while sorting samples, the cells are probably aggregating because of poor viability. We recommend diluting the cells and filtering the sample again using a 5-ml polystyrene round-bottom tube with a cell-strainer cap.
● TIMING
Staining panel #1: ~3 h–5 h 30 min
Steps 1–8, fresh samples: ~2 h or Steps 13–17, frozen samples: 30 min
Steps 35 and 36A, staining: 1 h 30 min
Steps 37 and 38, acquisition: ~1–2 h
Staining panel #2: ~3 h–5 h 30 min
Steps 1–8, fresh samples: ~2 h or Steps 13–17, frozen samples: 30 min
Steps 35 and 36A, staining: 1 h 30 min
Steps 37 and 38, acquisition: ~1–2 h
Staining panel #3: 3 h 30 min–7 h
Steps 1–8, fresh samples: ~2 h or Steps 13–17, frozen samples: 30 min
Steps 35 and 36B, staining: 1 h
Steps 37 and 39, sorting: ~2–4 h
Staining panel #4: 3 h–5 h 30 min + 8 d coculture
Steps 1–8, fresh samples: ~2 h or Steps 13–17, frozen samples: 30 min
Steps 18–26, culture of mouse MS-5 stromal cells: 8d
Steps 27–34, hematopoietic differentiation on MS-5 stromal cells: 8 d
Steps 35 and 36C, staining: 1 h 30 min
Steps 37 and 38, acquisition: ~1–2 h
Staining panel #5: ~3 h–5 h 30 min + 8 d coculture
Steps 1–8, fresh samples: ~2 h or Steps 13–17, frozen samples: 30 min
Box 1, CFSE staining: 30 min
Steps 18–26, culture of mouse MS-5 stromal cells: 8d
Steps 27–34, hematopoietic differentiation on MS-5 stromal cells: 8 d
Steps 35 and 36C, for staining: 1 h 30 min
Steps 37 and 38, acquisition: ~1–2 h
ANTICIPATED RESULTS
Typically, GMDPs, MDPs and CDPs are found in the Lin− CD34+ cells, which comprise only a small and variable fraction of human BM cells (2–4%) and CB cells (~1%): (i) GMDPs represent 5% (BM) and 10% (CB) of CD34+ cells; (ii) MDPs represent 0.5% (BM) and 0.5% (CB) of CD34+ cells; and (iii) CDPs represent 2% (BM) and 0.4% (CB) of CD34+ cells14. Pre-cDCs, which are found in the Lin− CD34− cells, represent 0.1% (BM) and 0.008% (CB) of CD45+ cells15. Therefore, DC progenitors are rare cells; some variability in the number of progenitors is expected from one donor to another, but progenitors are consistently found in all BM and CB specimens. Variability between subjects is particularly problematic for BM samples that are obtained from elderly donors, in which age-related changes may affect the composition and the phenotype of the cells.
As this protocol is based mainly on flow cytometry, it is crucial to verify the proper function and setup of the flow cytometer or cell sorter by running an appropriate cytometer procedure and making sure that laser delays are properly adjusted24. This step is often omitted and an improper setting will give inconsistent results.
To perform a full and comprehensive evaluation of BM and CB hematopoietic progenitors using multiparametric flow cytometry (panels #1 and #2), an ideal number of 2 × 106 (BM) or 4 × 106 (CB) cells are required. Therefore, a small fraction of BM or CB is used, allowing the majority of the sample to be used for other studies. This protocol provides an appropriate method for BM and CB analysis that could be applied to samples from healthy or abnormal donors. Moreover, the same procedure could be adapted to other tissues—that is, blood, lymph node or tonsil—in order to identify and sort pre-cDCs for further studies and applications. Of note, the low frequency of pre-cDCs in whole blood (~15 cells per ml of blood) dictates the use of several milliliters of blood for their quantification.
To ensure successful hematopoietic differentiation of DC progenitors in MS5+FSG culture, cell sorting should be performed under sterile conditions, and sorting parameters such as sheath fluid pressure should be optimized for viability of the cells. Ideally, a maximum of 104 CD34+ HSPCs or DC progenitors are plated on top of MS-5 stromal cells for 7 d. After 2 d of expansion, colonies can be observed under the microscope. The proportion of CD45+ cells substantially increases over 7 d. For early hematopoietic progenitors such as CD34+ HSPCs, the yield of cells can be increased a little bit by extending the culture; however, over 14 d expansion is associated with substantially lower output of CD45+ cells because of competition for cytokines and nutrients, as well as stromal cell death. The efficiency of the hematopoietic differentiation is going to depend on the sample itself, as variability between donors exists, and the number of cells that are initially cocultured on MS-5 stromal cells. After the cells are sufficiently expanded, further characterization can be performed using flow cytometry (panels #4 and #5).
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health (NIH) grant 1U19AI111825-01 (to M.C.N.), NIH grant AI13013, NIH/National Center for Advancing Translational Sciences (NCATS) grant UL1 TR000043, The Iris and Junming Le Foundation (to G.B.), The Empire State Stem Cell Fund through New York State Department of Health Contract no. C029562 (to K.L.) and NIH/National Institute of Allergy and Infectious Disease (NIAID) grant AI101251 (to K. Liu). M.C.N. is a Howard Hughes Medical Institute investigator.
Footnotes
AUTHOR CONTRIBUTIONS G.B., J.L., K.L. and M.C.N. conceived and developed the original strategy to identify the DC progenitors; G.B. modified the protocol and performed the experiments; G.B. and M.C.N. wrote the manuscript; and all authors reviewed the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Curr. Top. Microbiol. Immunol. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- 2.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 3.Manicassamy S, Pulendran B. Dendritic cell control of tolerogenic responses. Immunol. Rev. 2011;241:206–227. doi: 10.1111/j.1600-065X.2011.01015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 5.Jin JO, Zhang W, Du JY, Yu Q. BDCA1-positive dendritic cells (DCs) represent a unique human myeloid DC subset that induces innate and adaptive immune responses to Staphylococcus aureus infection. Infect. Immun. 2014;82:4466–4476. doi: 10.1128/IAI.01851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohn L, et al. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J. Exp. Med. 2013;210:1049–1063. doi: 10.1084/jem.20121251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachem A, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of murine CD8+ dendritic cells. J. Exp. Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crozat K, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8a+ dendritic cells. J. Exp. Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jongbloed SL, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulin LF, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8a+ dendritic cells. J. Exp. Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villadangos JA, Shortman K. Found in translation: the human equivalent of mouse CD8+ dendritic cells. J. Exp. Med. 2010;207:1131–1134. doi: 10.1084/jem.20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins SH, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 2015;212:385–99. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breton G, et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 2015;212:401–413. doi: 10.1084/jem.20141441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 17.Hao QL, et al. Identification of a novel, human multilymphoid progenitor in cord blood. Blood. 2001;97:3683–3690. doi: 10.1182/blood.v97.12.3683. [DOI] [PubMed] [Google Scholar]
- 18.Hoebeke I, et al. T-, B- and NK-lymphoid, but not myeloid cells arise from human CD34+CD38−CD7+ common lymphoid progenitors expressing lymphoid-specific genes. Leukemia. 2007;21:311–319. doi: 10.1038/sj.leu.2404488. [DOI] [PubMed] [Google Scholar]
- 19.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, et al. SCID-repopulating cell activity of human cord blood-derived CD34− cells assured by intra-bone marrow injection. Blood. 2003;101:2924–2931. doi: 10.1182/blood-2002-09-2782. [DOI] [PubMed] [Google Scholar]
- 21.Boissel N, et al. Defective blood dendritic cells in chronic myeloid leukemia correlate with high plasmatic VEGF and are not normalized by imatinib mesylate. Leukemia. 2004;18:1656–1661. doi: 10.1038/sj.leu.2403474. [DOI] [PubMed] [Google Scholar]
- 22.Bigley V, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J. Exp. Med. 2011;14:227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perfetto SP, et al. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat. Protoc. 2012;7:2067–2079. doi: 10.1038/nprot.2012.126. [DOI] [PubMed] [Google Scholar]