SUMMARY
Carbapenemases have become a significant mechanism for broad-spectrum β-lactam resistance in Enterobacteriaceae and other Gram-negative bacteria such as Pseudomonas and Acinetobacter spp. Intestinal carriage of carbapenemase-producing organisms (CPOs) is an important source of transmission. Isolation of carriers is one strategy that can be used to limit the spread of these bacteria. In this review, we critically examine the clinical performance, advantages, and disadvantages of methods available for the detection of intestinal carriage of CPOs. Culture-based methods (Centers for Disease Control and Prevention [CDC] protocols, chromogenic media, specialized agars, and double-disk synergy tests) for detecting carriage of CPOs are convenient due to their ready availability and low cost, but their limited sensitivity and long turnaround time may not always be optimal for infection control practices. Contemporary nucleic acid amplification techniques (NAATs) such as real-time PCR, hybridization assays, loop-mediated isothermal amplification (LAMP), or a combined culture and NAAT approach may provide fast results and/or added sensitivity and specificity compared with culture-based methods. Infection control practitioners and clinical microbiologists should be aware of the strengths and limitations of available methods to determine the most suitable approach for their medical facility to fit their infection control needs.
INTRODUCTION
At some point, almost all Enterobacteriaceae were susceptible to broad-spectrum β-lactam antibiotics, including β-lactam–β-lactamase inhibitor combinations, oxyimino-cephalosporins (e.g., ceftriaxone, ceftazidime, and cefotaxime), aztreonam, and carbapenems. Regrettably, two seminal events occurred in the past 30 years, which have had a major impact on the therapy of infectious diseases. In a manner analogous to the HIV epidemic and its human toll, the evolution of extended-spectrum β-lactamases (ESBLs) 3 decades ago significantly crippled the activity of oxyimino-cephalosporins and aztreonam, followed by the more recent appearance of carbapenemases in the clinic, which has limited the efficacy of all currently available β-lactams, causing a staggering economic and human burden (1). We have learned that increased colonization pressure from carbapenemase-producing organisms (CPOs) is linked to the development of infection (2), and gastrointestinal carriage of ESBL-producing Enterobacteriaceae leads to subsequent infection (3). Still today, after the initial report in 1983 of SHV-2 (the first ESBL reported), and despite significant advances in infection control and supportive care, infections caused by ESBL-producing Enterobacteriaceae exact an unacceptable mortality rate and add significantly to health care costs (4–6). The emergence of carbapenemases in the past 15 years has only added to the crisis caused by ESBL producers (7). The global impact of Klebsiella pneumoniae carbapenemase (KPC) and the New Delhi metallo-β-lactamase (NDM) created a worldwide fear that we are at the “end of the antibiotic era” (8, 9). The World Health Organization (WHO) has classified carbapenemase-producing Enterobacteriaceae (CPE) as one of the three greatest threats to human health (10). Surveys of the molecular epidemiology of carbapenemases, including KPC, OXA-48, VIM, IMP, and NDM producers, reveal that the dissemination of these carbapenemases is rapid and lasting. Authorities have advocated for local and regional screening programs, as available evidence shows that travelers are a major source of spread (11, 12). Furthermore, in settings where these organisms are endemic, transmission of ESBL-producing Enterobacteriaceae between health care facilities creates a significant challenge for controlling the spread of resistance (13). The number of CPE cases in community hospitals in the Southwestern United States has increased 5-fold in the last few years (14).
Current Status of Carbapenemases
Carbapenemases are present among all four classes of β-lactamases (Table 1) (15–17). A rare class C β-lactamase, CMY-10, also demonstrates weak “carbapenemase activity,” but its clinical significance is unclear (17, 18). Additionally, CPOs are commonly resistant to multiple drug classes, such as aminoglycosides, quinolones, tetracyclines, and folate inhibitors, due to additional types of resistance genes carried by the organisms (19, 20). To provide the appropriate background for evaluating the detection methods discussed here, we review the major carbapenemases that are threatening our β-lactam arsenal.
TABLE 1.
Carbapenemases and selected characteristicsa
| Molecular class | Representative β-lactamase | Characteristic(s) | Inhibitor(s) | Enzyme currently found in areas of endemicity | Area(s) of endemicity |
|---|---|---|---|---|---|
| A | KPC, GES, SMC | Serine β-lactamase, plasmid encoded | Boronic acid derivatives | KPC | North America, Greece, Italy, Poland, Colombia, Argentina, Israel, China |
| GES-5 | Brazil | ||||
| B | NDM, VIM, IMP, GIM-1, SPM | Metallo-β-lactamase, zinc requiring, plasmid encoded/chromosomal | EDTA, dipicolinic acid | NDM | Indian subcontinent, Kenya, China |
| VIM | Indian subcontinent, Greece, Italy, southern France, Japan, Lebanon, Brazil, Portugal, Ireland, UK, Germany, Poland | ||||
| IMP | Indian subcontinent, Greece, Japan, China | ||||
| C | CMY-10 | Serine β-lactamase, cephalosporinases, mobile or chromosomal, uncommon | Cloxacillin, boronic acid derivatives | AmpC | Worldwide |
| D | OXA-48, OXA-181, OXA-204, OXA-162, OXA-23, OXA-24 | Serine β-lactamases, weak activity of those that are carbapenemases, plasmid encoded | No specific inhibitors available | OXA-48 | France, Belgium, Canada, South Africa, Middle East, Turkey, northern Africa, Switzerland, Germany, Lebanon, Israel, Morocco |
Class A carbapenemases.
One of the most common mechanisms of carbapenem resistance among class A enzymes is the production of KPC β-lactamases. KPCs were initially detected in a clinical isolate in 1996 in North Carolina; since then, 19 variants have been discovered (21–24). KPC has been found in a variety of Enterobacteriaceae, including Klebsiella spp., Escherichia coli, Enterobacter spp., Citrobacter spp., Morganella spp., Serratia marcescens (25–29), Raoultella spp. (30), Kluyvera (31), and Salmonella (32), and in non-Enterobacteriaceae such as Aeromonas (33), Pseudomonas, and Acinetobacter baumannii (34).
Attributable and crude mortality rates for infections caused by bacteria harboring KPCs are higher than those for patients with non-KPC-producing isolates (35); the reason for this increased mortality is still enigmatic. Epidemiological studies suggest that KPC-producing K. pneumoniae isolates belonging to sequence type 258 (ST258) are of two distinct clones and that the clinical behavior of isolates bearing blaKPC-2 is different from that of isolates carrying blaKPC-3. Molecular differences between the two clones include aminoglycoside resistance and the ability to form biofilms (36, 37). The molecular reason for this difference in clinical behavior is not yet understood. The prevalences of KPC-producing bacteria vary widely. In one surveillance study, 37% of patients in an intensive care unit (ICU) carried blaKPC (38). Other studies place its prevalence at between 0 and 5%, depending on the population being surveyed (39, 40).
Some areas of Europe (Greece, Italy, and Poland), South America (Colombia and Argentina), the Middle East (Israel), and North America are areas where KPC is endemic. Recently, cases and localized outbreaks have been linked to importation from areas of endemicity (22, 41). In addition, long-term-care facilities (LTCFs) are rapidly becoming reservoirs for KPC producers (41). Other class A carbapenemases are important in some specific locales, such as GES-5 in Brazil, where it constitutes the main carbapenemase in Enterobacteriaceae (22). SME carbapenemases, also belonging to class A and associated with Serratia marcescens, are quite rare.
Class D carbapenemases.
Another important carbapenemase in Enterobacteriaceae is a class D β-lactamase, OXA-48. This β-lactamase, sometimes referred to as the “phantom menace,” was initially identified in a Turkish patient in 2001 (42–44). For the next 5 years, OXA-48 was not isolated from any other country. In 2008, OXA-48 spread outside Turkey and became prevalent in clinical isolates from Continental Europe, the Middle East, and northern Africa (45, 46). Since then, outbreaks throughout Europe have been reported (45, 47). Most recently, OXA-48 was detected in the United States, Canada, and South Africa (20, 48–50). Many of these reports involve patients previously treated in Middle Eastern and North African countries (51). Nonetheless, an early outbreak of OXA-48-producing K. pneumoniae in England was not linked to known regions of endemicity (52). More concerning, however, was a retrospective analysis that uncovered an outbreak of OXA-48-producing Enterobacteriaceae in a Dutch hospital that had been ongoing for 2 years (53). OXA-48 has disseminated to a wide variety of Enterobacteriaceae species, including Klebsiella spp., E. coli, Citrobacter spp., Serratia marcescens (54–56), Enterobacter spp., Morganella morganii (55), Providencia stuartii (57), Raoultella planticola (56), and Salmonella enterica (51).
OXA-48 is contained in a 61.8-kb self-conjugating IncL plasmid, which likely contributes to its ability to spread in Enterobacteriaceae (22, 58, 59). Other OXA-48-like enzymes with carbapenemase activity in Enterobacteriaceae that either have caused or have the potential to cause outbreaks include OXA-181, OXA-204, OXA-232, and OXA-162 (22, 43, 60–62). Other class D carbapenemases of clinical importance are OXA-23 and OXA-24/40; these carbapenemases are found mainly in Acinetobacter baumannii (63). Recently, some OXA-type carbapenemases have been reclassified based on their hydrolytic activity. To illustrate, once thought to be a carbapenemase, the kinetic profile of OXA-163 resembles more an ESBL than a carbapenemase (62).
Class B carbapenemases.
The class B metallo-β-lactamases (MBLs) hydrolyze a broad range of β-lactams, including carbapenems (18). The most widespread MBLs include the NDM, VIM, and IMP family enzymes. Of the MBLs, NDM-1 has emerged as a major cause of concern due to its widespread dissemination (64). NDM-1 was initially detected in a patient of Indian origin in Sweden in 2007 (65). NDM-1 was subsequently found to be widespread in the Indian subcontinent, including in environmental samples (66), and has now been reported in more than 15 countries (67). In the United Kingdom, 52% of 101 patients with NDM-producing isolates collected from 2008 to 2013 reported health care exposure or travel to the Indian subcontinent (68). NDM has spread between different bacterial species, including Enterobacter cloacae, K. pneumoniae, and Escherichia coli (69).
Horizontal spread of NDM has also been described in the clinical setting; in a recent study, four neonates from India acquired an NDM-1-producing E. coli isolate from the environment and developed sepsis (70). Clonal spread of NDM-1-producing isolates has been documented in some regions of India, while spread elsewhere, including to the United Kingdom, likely happened due to a transfer of plasmids (71).
Equally important, IMP- and VIM-producing bacteria have also been found in the United States, Europe (mostly Greece, Italy, and southern France), the Middle East, the Indian subcontinent, Japan, and China (22, 72–74). Outbreaks have occurred throughout the world, as these MBLs spread as part of complicated integrons (42). To illustrate, a recent surveillance study performed in northeastern Ohio uncovered a clinical isolate of Pseudomonas aeruginosa with blaVIM-2 in a class I integron that was proximal to a Salmonella genomic island (SGI), suggesting recombination between these two bacteria. A detailed analysis of this genetic locus showed multiple resistance and transposing elements that likely resulted in the successful dissemination of this isolate (75).
MECHANISMS OF CARBAPENEM RESISTANCE
Resistance to carbapenems can be mediated by different mechanisms; these include porin mutations, upregulation of efflux pumps, changes in penicillin binding proteins (PBPs), and production of carbapenemases (76–78). A significant subset of carbapenemase genes are harbored in readily transmissible plasmids. These plasmids, in some circumstances, can be shared between Enterobacteriaceae and non-Enterobacteriaceae. While other mechanisms of resistance are also genetically encoded, their transmission is not as frequently observed as for carbapenemase genes and therefore are of lesser concern.
In this treatise, we generally refer to carbapenem-resistant organisms (CROs) as bacteria that are resistant to imipenem, meropenem, doripenem, or ertapenem. We particularly focus on Gram-negative CROs. These can be divided in Enterobacteriaceae and non-Enterobacteriaceae. Carbapenem-resistant Enterobacteriaceae are frequently referred to as CRE. Organisms that are carbapenem resistant due to the production of a carbapenemase are referred as carbapenemase-producing organisms (CPOs), and when the bacteria are Enterobacteriaceae, we refer to them as carbapenemase-producing Enterobacteriaceae (CPE). In addition, there are some bacteria that produce carbapenemases even though their MICs for carbapenems do not reach the resistance breakpoint. Given that carbapenemase genes are usually transmissible via plasmids, we argue that they should be targeted for screening, and we include them as CPOs or CPE. It must be noted that some non-Enterobacteriaceae CPOs such as Burkholderia spp. and Stenotrophomonas maltophilia carry chromosomally encoded carbapenemases. As such, the chromosomally encoded carbapenemases are unlikely to be transmitted to other bacteria. When we refer to CPOs in this review, we focus on all Enterobacteriaceae and the non-Enterobacteriaceae that are known to carry carbapenemase-encoding plasmids, even when their MIC increase does not reach the resistance breakpoint.
INTESTINAL CARRIAGE OF CPOs
As noted above, CPOs have emerged as significant health care-associated pathogens worldwide (38, 79). While most studies refer to CPE, we contend that similar conclusions can be applied to CPOs, encompassing both Enterobacteriaceae and nonfermenting Gram-negative bacteria. Furthermore, since carbapenemases are transferred via plasmids, both Enterobacteriaceae and non-Enterobacteriaceae are capable of serving as reservoirs and vectors. Intestinal carriage serves as a reservoir of CPE and can promote cross-transmission in health care settings (80). Thus, infection control programs directed at detecting intestinal carriage are essential tools to limit the spread of these pathogens.
Several examples highlight the importance of detection of intestinal carriage for the effective control of infections due to CPOs. A study in New York documented a significant decrease in the carriage rate 1 year after an infection control program in an ICU was implemented (81). The program involved screening for intestinal carriage of carbapenem-resistant K. pneumoniae and Acinetobacter baumannii with culture of rectal swabs (BBL CultureSwab Plus; Becton-Dickinson) and isolating patients while results were pending or if they were positive. Isolation was carried out in the rooms at the far end of an ICU, where rooms were divided only by curtains. The program also involved extensive cleaning with isopropanol and a quaternary ammonium compound, which included closing the unit for 2 days. The lack of a quick screening test was a limiting factor for the success of this program. Nonetheless, those investigators were able to reduce the mean number of new KPC-producing K. pneumoniae cases from 9.7 to 3.7 per 1,000 patient-days. In another study, Enfield et al. were successful in decreasing the CPE incidence in a surgical ICU from 7.77 cases per 1,000 patient-days to 1.22 cases per 1,000 patient-days by using enhanced infection control measures and increased surveillance by implementing a PCR-based assay (82). Two outbreaks of KPC-producing organisms were successfully controlled by using a “bundle approach,” of which screening for CPE carriage is an integral part (83, 84). Schwaber et al. reported on a country-wide mandatory program involving physical isolation and dedicated nursing staff in Israel that was able to significantly decrease the incidence of KPC-producing isolates (85). Although screening for asymptomatic carriage was not part of the program, this effort involved a very broad isolation policy that relied on careful tracking of known cases throughout the health care system. In addition to these real-life examples, a mathematical model also validates the usefulness of screening followed by patient isolation to control CPOs (86).
Failure of Carbapenem Breakpoints To Detect All CPOs
Detection of CPOs in the clinical microbiology laboratory is challenging because interpretation of routine susceptibility test results may fail to flag an isolate as a potential CPO (14, 87–89). Although the presence of a carbapenemase confers some resistance, the increase in the MICs due to the β-lactamase may not be enough to consider the isolate resistant to a carbapenem given the defined cutoff values for interpretation of resistance (90). Despite the changes made by the Clinical and Laboratory Standards Institute (CLSI) to the carbapenem interpretative criteria for Enterobacteriaceae in June 2010, which lowered the MIC values for isolates that were considered “nonsusceptible” (from ≤4 μg/ml to ≤1 μg/ml for meropenem) to detect more CPE than under previous guidelines, there are some isolates that still escape detection (90). Recently, both the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the CLSI have proposed not testing for resistance mechanisms in clinical isolates, arguing that the lower breakpoints should suffice for treatment purposes (Table 2) (91, 92).
TABLE 2.
Clinical breakpoints for carbapenems according to CLSI and EUCAST guidelinesa
| Carbapenem | Susceptibility breakpoint (μg/ml) according to: |
|||
|---|---|---|---|---|
| CLSI document M100-S20, 2010 (196) | CLSI document M100-S20U, 2010 (197) | CLSI document M100-S22, 2012 (198) | EUCAST, 2009–2014 | |
| Doripenem | Not defined | ≤1 | ≤1 | ≤1 |
| Ertapenem | ≤2 | ≤0.25 | ≤0.5 | ≤0.5 |
| Meropenem | ≤4 | ≤1 | ≤1 | ≤2 |
| Imipenem | ≤4 | ≤1 | ≤1 | ≤2 |
An intermediate result is interpreted as an MIC that is 1 dilution higher, and a resistant result is interpreted as an MIC ≥2 dilutions higher, except for the EUCAST interpretation of ertapenem results, where an organism with an MIC of >1 μg/ml is considered resistant. CLSI documents M100-S23 (199) and M100-S24 (200) (2013 and 2014) do not change the interpretative criteria for carbapenems. CLSI document M100-S22 (2012) (198) changed interpretative criteria only for ertapenem. The criteria for other carbapenems were not changed. Doripenem was not included in the 2009 and 2010 editions of CLSI document M100.
We must emphasize that clinical breakpoints are meant for implementation in the care of patients and are not designed for epidemiological surveillance. In any case, uniform consensus on this issue does not exist; we agree with Livermore et al. and advocate for testing for carbapenemase resistance genes for infection control monitoring as well as for routine microbiological diagnosis (93). The rationale for this assertion is that although increased MICs against carbapenems may suggest the presence of a CPO, clinical experience demonstrates that MICs will not always reveal the presence of carbapenemases. The EUCAST proposed the use of epidemiological breakpoints followed by phenotypic confirmation by inhibition disks or the Carba NP test for this purpose (94). All of these changes reflect the notion that to prevent the spread of resistance, it is necessary to prevent the transmission of not only isolates that are phenotypically resistant but also those that carry transmissible elements that may spread to susceptible bacteria to confer resistance under the right conditions.
Carbapenemases, when not accompanied by other β-lactamases, may confer a low level of resistance to carbapenems (even MICs of ≤0.5 μg/ml) that does not become evident until they are combined with another resistance mechanism, such as the production of an ESBL or acquired AmpC (95, 96), porin mutations (97, 98), or changes in porin expression (99). Conversely, these changes may increase carbapenem MICs in the absence of carbapenemases. OXA-48 is particularly known for consistently failing to be detected if not accompanied by another broad-spectrum β-lactamase (100). This was demonstrated in the unrecognized Dutch outbreak described above (53).
Reports show that automated susceptibility systems are also not reliable for the detection of carbapenemase-producing K. pneumoniae isolates (101). According the interpretative criteria for meropenem from the 2005 CLSI document M100-S15 (which corresponds to document M100-S22, 2012, in Table 2), rates of nonsusceptible isolates in a panel of confirmed KPC producers ranged from 93% with the Microscan system (Beckman Coulter, Brea, CA, USA) to 20% with Sensititre AutoReader (Thermo Scientific, Waltham, MA, USA), compared with 100% for broth microdilution and disk diffusion (102). For KPC-producing non-Klebsiella isolates, the rate of false-negative results may be higher, although there has been some improvement using the revised CLSI breakpoints (88). The use of stricter criteria and expert rules for automated systems has increased the sensitivity of CPE detection but with a significant decline in specificity (103). A study comparing disk diffusion, Etest, and Vitek2 (bioMérieux, Marcy l'Etoile, France) methods using previous CLSI and EUCAST breakpoints for meropenem, imipenem, or ertapenem found multiple discrepancies with KPC, ESBL, and MBL producers. However, the Vitek2 system, when using meropenem as a reporter substrate, successfully detected all CPE producers (87).
On an operational basis, ertapenem and meropenem are proposed to be the most suitable antibiotics for screening of carbapenemase producers (89). Anderson et al. found that, depending on the method used, 0 to 6% of KPC-producing isolates were susceptible to ertapenem according to the former CLSI ertapenem resistance breakpoint of ≥8 μg/ml. After the breakpoint was decreased to ≤2 μg/ml (which is higher than the current breakpoint of 0.5 μg/ml), almost all tested methods were able to detect 100% of the KPC producers. Interestingly, the Vitek2 platform, when using ertapenem as an indicator, still failed to detect 6% of these isolates (89). The EUCAST guidelines reflect these issues, suggesting that meropenem (with a cutoff of 0.125 μg/ml) is the antibiotic with the best balance between sensitivity and specificity while noting that ertapenem is the most sensitive but lacks specificity (94). Faropenem, a penem antibiotic, has also been proposed as an alternative to carbapenems for the detection of carbapenemases (104, 105). Faropenem showed 99% sensitivity and 94% specificity when tested against a known panel of 166 PCR-confirmed isolates of carbapenemase-producing Enterobacteriaceae (although OXA-48-producing isolates were underrepresented) and 82 negative controls. Another study compared a panel of 62 PCR-confirmed KPC-producing Enterobacteriaceae and 73 producers of other β-lactamases, showing a nonoverlapping inhibitory zone around a 5-μg faropenem disk between KPC producers and nonproducers (104).
Tests for Carbapenemase Activity in Isolated Cultures
Tests that detect carbapenemase activity in isolated cultures within a short time period can be used to rapidly determine if a clinical isolate is a CPO. These tests are generally not suited for direct testing of nonsterile specimens without prior isolation or enrichment steps, so they would not be used to screen fecal specimens or perirectal swabs directly. However, they can be employed as confirmatory assays when using culture-based screening.
The modified Hodge test (mHT) was the initial screening test recommended for carbapenemase production (89). However, mHT lacks specificity and may produce false-positive results for bacteria with complex ESBL or AmpC (both plasmid and overexpressed chromosomal enzymes) backgrounds combined with porin mutations/loss (106, 107). For the mHT, it must be noted that this test should be performed with either meropenem or ertapenem, as it is known to perform poorly when imipenem is used as a substrate (92). When focusing on KPC enzymes, the specificity of this test can be increased by using an EDTA disk, as described by Yan et al. (108); however, the most common reason for false-positive results (i.e., AmpC hyperproduction) is not addressed with this modification. The mHT is also unable to distinguish between carbapenemases, and it lacks sensitivity for some carbapenemases such as NDM (particularly low-level producers), some members of the OXA family, and SME (107, 109).
Synergy testing with inhibitors can be used to differentiate MBLs from other enzymes by inhibiting class A, C, and D enzymes. The phenylboronic acid double-disk synergy test (PBA-DDST) with either meropenem or ertapenem was able to successfully screen for KPC β-lactamase in a collection of clinical specimens (110). Pournaras et al. evaluated PBA and EDTA with meropenem in disks in a sample of bacterial colonies isolated from 189 rectal swabs where 97 were positive for a carbapenemase (KPC and VIM) and showed excellent sensitivity and specificity (111). Doi et al. also showed that inhibition by PBA could be used to differentiate KPC from other β-lactamases (112).
Another proposed improvement to disk-based testing that allows differentiation between the different β-lactamase classes is the use of avibactam (formerly NXL104) disks (113, 114). Class C enzymes can also be identified with similar inhibition tests (115, 116). Further improvement of these tests allows the identification of concurrent mechanisms, such as KPC with a MBL, such as the one suggested by Miriagou et al. (117). In that study, PBA improved MBL inhibition, allowing fewer misclassifications of VIM and KPC than with the use of PBA. These disk tests, however, require pure cultures, making them inappropriate for screening of the lower gastrointestinal tract of patients. Nevertheless, these assays remain a low-cost, low-technology option.
The Carba NP test, the Blue-Carba test, and the Rapid Carb test are based on the detection of carbapenemase activity. The Carba NP test (bioMérieux, France) detects a change in pH that is coupled to the hydrolysis of imipenem. The testing procedure consists of cell lysis followed by incubation of the enzymatic lysate with imipenem and phenol red for up to 2 h (118–120). Testing of Enterobacteriaceae showed that Carba NP was able to detect all tested isolates from a worldwide collection of bacteria that produced class A (KPC, NMC, SME, and GES), class B (IMI, NDM, VIM, and IMP), and class D (OXA-48 or OXA-181) enzymes (119). In this report, false-positive results were not detected. The Carba NP test has also been used directly on blood culture bottles spiked with an array of class A, B, and D carbapenemases, where this assay demonstrated a sensitivity and a specificity of 97.9% and 100%, respectively (121). However, in another study, the Carba NP test produced noninterpretable results for testing of isolates grown on MacConkey or Drigalski agar (120), suggesting that these media may affect the performance of the test. Also, the use of the Carba NP test on A. baumannii may require some modifications (122). In any case, the Carba NP test is one of the recommended tests for confirmation of carbapenemase production in pure isolates by the CLSI and EUCAST (92, 94).
The Carba NP test can be performed in most microbiology laboratories, with no additional equipment. It can be used on any isolate with suspected carbapenemase activity. The main advantage over agar screening and molecular methods is the broad target range, as the test result will be positive as long as there is enough carbapenemase, regardless of its class. However, there is some concern regarding the lack of sensitivity for certain class D carbapenemases (e.g., OXA-48, OXA-58, and OXA-181) (123, 124). A possible drawback compared to molecular methods (and disk inhibition testing) is its inability to differentiate between enzymes, although one report suggests that this can be achieved with some modifications to the Carba NP test (125). The Carba NP test has also been compared to an alternative colorimetric test for carbapenemase activity, the Rapid Carb Screen (Rosco Diagnostics, Denmark), showing a similar sensitivity (97 and 98%) but superior specificity (100 and 83%) when tested against a panel of 66 Enterobacteriaceae isolates carrying class A, B, or D enzymes and 69 non-carbapenemase producers (126). Another test that is similar to the Carba NP test, the Blue-Carba test (Rapid Carba Screen; Rosco Diagnostics), uses a different indicator and a simplified protocol. The Blue-Carba test has a higher reported sensitivity, including for OXA-type carbapenemases (127). An added advantage of the Blue-Carba test is a faster turnaround time than that of the Carba NP test, as there is no need to extract the β-lactamase.
Spectrometry has also been used to detect CPOs. These tests include UV spectrophotometry and mass spectrometry using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). UV spectrophotometry involves the detection of hydrolyzed imipenem by a cell lysate (128). This method was found to be 100% sensitive and specific for detecting a wide array of class A, B, and D enzymes in Enterobacteriaceae (129). The clinical application of this method is still challenging due to the technical expertise and equipment required to perform it.
MALDI-TOF MS can detect carbapenemases by comparing the proportions of hydrolyzed and intact imipenem in a centrifuged cell sample (130). This approach was able to detect 72% of carbapenemase-producing isolates directly from positive blood culture vials (131). Although still a research application, this method may become attractive as MALDI-TOF MS becomes more common in the microbiology laboratory, but this method is suboptimal for the detection of small molecules, such as carbapenems and their degradation products. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is more suitable for this task and has also been used successfully to detect carbapenemase activity from cultures (132). These methods have the potential to differentiate between classes of β-lactamases by using inhibitors such as EDTA (133). It must be noted that imipenem undergoes spontaneous hydrolysis in basic buffers (134) and that a negative control should always be included when using any of these methods. At this time, a mass spectrometry method that is as sensitive and easily implemented as agar-based or PCR-based screens is not yet present.
Screening Methods To Detect Fecal Carriage of CPOs
Screening tests to detect CPOs in stool present three major challenges: rapid detection, detection of isolates with low-level carbapenem resistance, and detection of proportionally low numbers of CPOs. Infection control programs rely on contact isolation for patients who test positive, which also must be in place while waiting for the result. A “good screening test” must minimize turnaround time, maximize sensitivity, preserve reasonable specificity, detect multiple types of carbapenemases, and be cost-effective. Detection of low-level resistance is important because it may already signify the presence of a genetic trait (such as blaKPC) (90) that may spread to other bacteria through horizontal transfer, where it could result in carbapenem resistance in new bacterial strains (135). Finally, since the main reservoir is the intestinal tract, the bacteria of concern may represent just a small proportion of the overall bacterial load. Therefore, the inoculum of CPOs on a surveillance swab may be below the limit of detection (LOD). It is also worth mentioning that non-Enterobacteriaceae Gram-negative bacteria may also harbor carbapenemases, although screening protocols based on culture methods try to exclude them. For instance, U.S. Centers for Disease Control and Prevention (CDC) protocols and the suggested interpretation for the Supercarba agar test suggest that only lactose-fermenting colonies should be reported (see Fig. 1). Manufacturers of chromogenic media also endorse the reporting of colonies with certain appearances that correspond to lactose fermenters. Bacteria other than lactose-fermenting Enterobacteriaceae can be detected with appropriate culture methods, and molecular screening tests will yield a positive result if a carbapenemase is present, regardless of the host organism. Inclusion of bacteria other than lactose fermenters in a screening program is important, as they can also transmit resistance elements to or within the Enterobacteriaceae, as has been previously suggested (75).
FIG 1.
Appearances of different Enterobacteriaceae on chromID Carba and Supercarba media. (Left) chromID Carba plate. Red colonies represent K. pneumoniae, blue colonies represent E. coli, and yellow colonies represent Pseudomonas aeruginosa. (Right) Supercarba medium composite picture. The top half shows K. pneumoniae (yellow colonies due to lactose fermentation). The bottom half shows Pseudomonas aeruginosa (black/dark green colonies with no lactose fermentation).
A summary of tests is provided in Table 3. As shown, the cost, labor intensity, and turnaround time vary by assay. Mathers et al. reported that the annual costs of a surveillance program for a hospital containing 708 acute-care beds and 40 long-term beds with weekly screening and a CPE prevalence of 2.7% were about $225,000 for a qPCR (quantitative real-time PCR) assay and $23,000 for the CDC screening culture method (136). Although Mathers et al. accounted for the cost of decreased specificity, the cost of decreased sensitivity is much more difficult to calculate. For instance, a false-positive result (product of low specificity) would result in further follow-up testing; however, a false-negative result (product of low sensitivity) may result in the spread of the CPO, potentially adding very significant costs for the hospital to care for infected patients, while instead, it would appear to decrease the cost of the screening program. The apparent difference between costs of methods can translate into many thousands of dollars per year for a hospital performing routine screening in large volumes. Added to the cost of screening is the cost of isolation. A 2014 Canadian study estimated a cost of Can$925 (approximately US$740) per non-ICU patient when a patient is isolated for 3 days while awaiting results (137).
TABLE 3.
Characteristics and approximate costs of screening methods to identify fecal carriage of CPE
| Method (reference) | Description | Turnaround time for positive or preliminary positive result (h) | Price (US$)a |
|---|---|---|---|
| CDC protocol (138) | Broth enrichment of rectal swab in ertapenem medium followed by subculture on MacConkey agar with carbapenem disk, followed by identification of suspect isolates | 48–72e | Negative test result, 1–2b; positive test result, 2–6b |
| Supercarba (149) | Direct plating of rectal swab in selective medium | 24–48e | 1c |
| Chromogenic medium | Direct plating of rectal swab in selective medium with chromogenic molecule | 24–48e | 4–7 |
| Real-time PCRd (in-house methods) | DNA extraction followed by PCR and probe-based detection | 2–5 | 10–30b |
| Commercial PCR assayd | DNA extraction followed by PCR and probe-based detection. | 2–3 possible | 30–60b |
U.S. dollars as of 2015.
Based on cost data by Mathers et al. (136).
Supercarba medium has been patented. The cost is that of raw materials; this medium is not available to many laboratories.
The cost of the PCR assay may increase with increased numbers of targets.
Confirmation testing might include singleplex PCR, multiplex PCR, Carba NP, Blue-Carba, or identification and susceptibility testing. The cost may range from an additional $2 to $50, and the turnaround time may range from an additional 2 to 24 h for confirmatory testing, depending on the methods chosen by the laboratory. Hospital epidemiology can act on negative results and preliminary positive results, pending confirmation. Negative NAAT results likely do not require confirmatory testing, but positive results may require confirmation, depending on the false-positive rate of the assay.
Culture-based methods.
Culture-based testing is easier to implement, as the necessary equipment and knowledge are already present in the routine microbiology laboratory. These tests also have the potential to detect reduced susceptibility to carbapenems caused by newly emerging mechanisms as long as the mechanism is able to achieve at least a moderate level of resistance.
The CDC screening method addresses, with significant limitations, the need for the detection of low-level resistance (MIC < 2 mg/liter) and the ability to detect low loads of resistant bacteria. This method consists of an enrichment phase where a rectal swab is inoculated into 5 ml of Trypticase soy broth (TSB) in which a disk impregnated with 10 μg of ertapenem or meropenem has been immersed and incubated for 24 h. This broth is then subcultured onto MacConkey agar, where only lactose fermenters are selected. The CDC notes that many laboratories add a meropenem or ertapenem disk to this agar. A limitation of this test is that further testing is needed to determine the species and antimicrobial susceptibility of isolates growing on the agar (138). Furthermore, bacteria other than lactose fermenters that can harbor carbapenemases are routinely missed. Given the increased length of time needed for detection when using methods such as the CDC method, selective agars (see below) have been developed to optimize detection while obtaining results in a shorter time span. More important is that the CDC method will fail to detect the presence of bacteria with low-level resistance unless these bacteria are present in a large inoculum and without competition with other CROs, conditions that are unlikely to happen. Furthermore, small inocula of fully resistant CPOs can be missed if there is a large inoculum of bacteria that have low-level carbapenem resistance through mechanisms other than carbapenemases. Although the CDC broth enrichment method was meant to increase sensitivity, recent reports demonstrate that some of the selective agar methods have a performance that is superior or at least comparable to that of the CDC method, so the delay of an overnight enrichment is not necessary (139–141).
Specialized solid media aim to simplify the detection of CPE. Chromogenic media incorporate chromogenic enzyme substrates (mainly glycosides) that release a pigment when hydrolyzed by bacterial enzymes (142). Antibiotics added to the media make them selective for a particular resistance trait. Chromogenic media have been compared regarding their limit of detection of CPE with different inocula when used for stool screening (80, 139, 143–146). At this time, the currently available media have not been cleared by the U.S. Food and Drug Administration (FDA).
Available chromogenic media that may be used for the detection of carbapenemases include CHROMagar KPC (CHROMagar, France), HardyChrom (Hardy Diagnostics, CA, USA), chromID Carba (bioMérieux, France) (Fig. 1), chromID ESBL (bioMérieux, France), chromID OXA-48 (bioMérieux, France), Colorex KPC (Biomed Diagnostics, OR, USA), RambaChrom KPC (Gibson Bioscience, USA), SpectraCRE (Thermo Diagnostics, USA), and Brilliance CRE (Thermo Diagnostics, USA). Colorex KPC medium consists of medium commercially prepared from dry CHROMagar reagents.
Some of these media are designed to target KPC producers and have markedly decreased sensitivity for mechanisms based on other enzymes, particularly OXA-48 (143, 147). This is specifically addressed with a medium designed for the detection of OXA-48 producers, chromID OXA-48.
Table 4 shows the performance characteristics of different chromogenic media when tested with pure cultures. The specificity varies depending on the type of negative controls used (clinical specimens or known non-carbapenemase-producing but carbapenem-resistant isolates). In addition, although not shown in Table 4, all testing methods had slightly but consistently lower sensitivities for VIM β-lactamases than for other class B β-lactamases (80, 139, 143–146). This may be due to the inclusion of isolates that contained plasmids carrying at most another β-lactamase, rather than isolates with more complex backgrounds that have now become prevalent (148). However, this likely does not hold true for bacteria harboring VIM-containing plasmids that also carry an ESBL or another carbapenemase.
TABLE 4.
Use of known resistant isolates to test performance of CPE screening methodsa
| Method | Overall sensitivity (%) | Sensitivity (%) by β-lactamase class (no. of isolates tested) |
Specificity (%) (no. of negative isolates tested | Type(s) of isolates | Reference | ||
|---|---|---|---|---|---|---|---|
| Class A | Class B | Class D | |||||
| Supercarba | 95.6 | 100 (18) | 90 (52) | 100 (44) | 82.2 (62) | CLI | 149 |
| chromID ESBL | 87.7 | 100 (18) | 98 (52) | 70 (44) | 24.2 (62) | ||
| CHROMagar KPC | 40.3 | 66.7 (18) | 55.8 (52) | 13.6 (44) | 85.5 (62) | ||
| Supercarba | 96.5 | 100 (20) | 92 (51) | 100 (43) | 60.7 (28) | CLI | 143 |
| CHROMagar KPC | 43 | 70 (20) | 58.8 (51) | 11.6 (43) | 67.8 (28) | ||
| Brilliance CRE | 76.3 | 85 (20) | 78.4 (51) | 69.8 (43) | 57.1 (28) | ||
| Brilliance CRE | 86 | 100 (17) | 72 (25) | 88 (58) | 40 (77) | CLI | 147 |
| Colorex KPC | 48 | 100 (17) | 52 (25) | 31 (58) | 39 (77) | ||
| Supercarba | 97 | 100 (17) | 88 (25) | 100 (58) | 35 (77) | ||
| Brilliance CRE | 78 | 83 (12) | 79 (103) | 67 (15) | 66 (70) | CLI | 144 |
| chromID Carba | 91 | 100 (12) | 93 (103) | 67 (15) | 89 (70) | ||
| chromID ESBL | 96 | 100 (12) | 98 (103) | 80 (15) | 19 (70) | ||
| Colorex KPC | 56 | 83 (12) | 52 (103) | 60 (15) | 77 (70) | ||
| CDC protocol for ertapenem | 78 | 83 (12) | 80 (103) | 73 (15) | 69 (70) | ||
| CDC protocol for meropenem | 47 | 67 (12) | 46 (103) | 40 (15) | 79 (70) | ||
| Brilliance CRE | 94 | 100 (36) | 94 (34) | 84 (25) | 71 (160) | CLI, CCI | 146 |
| mHT | 100 | 100 (18) | ND (0) | ND (0) | 96.7 (32) | CCI | 110 |
| RambaChrom KPC | 95 | 95 (18) | ND (0) | ND (0) | 77.1 (32) | ||
| Mero-PBA-DDST | 100 | 100 (18) | ND (0) | ND (0) | 100 (32) | ||
| Erta-PBA-DDST | 100 | 100 (18) | ND (0) | ND (0) | 91.4 (32) | ||
CCI (characterized clinical isolate) is an isolate originating from a clinical specimen and later characterized in the laboratory. CLI (characterized laboratory isolate) is an isolate retrieved from a laboratory source. It may have originated from a clinical specimen but may have been modified to express certain characteristics in the laboratory. ND, not determined; mHT, modified Hodge test; PBA-DDST, phenylboronic acid double-disk synergy test; Mero, meropenem; Erta, ertapenem.
Supercarba agar (Fig. 1) is another specialized medium that incorporates the use of ertapenem (0.5 mg/liter) in addition to cloxacillin in a zinc-supplemented Drigalski lactose agar (149). Ertapenem will select for carbapenem resistance, and cloxacillin is added to inhibit the growth of AmpC producers such as Serratia and Enterobacter species, while zinc enhances the activity of MBLs (149). Different studies have shown a sensitivity of ∼96% and a specificity of 60%. These numbers are similar to those obtained with chromogenic media (143, 149). Those authors recommend the selection of only lactose-fermenting bacteria, limiting the ability of this method to detect carbapenemases in bacteria other than lactose fermenters. Another disadvantage is that the shelf life of the medium is limited to 7 days, a significant obstacle in any routine clinical laboratory (143).
Relative performance of culture methods.
Studies describing different methods for screening of CPE are difficult to compare, and each study has its own limitations and particular variations. Some studies have addressed the detection limit of different commercial assays by using previously characterized CPE isolates. These isolates, however, may not be representative of the population at a specific hospital, and clinical performance in actual practice may vary due to the prevalence of different β-lactamases in different institutions. Meanwhile, other studies have compared the performances of tests in a particular setting, such as hospitals, where there is a particular distribution of resistance mechanisms within the bacterial population. It is difficult to extrapolate the performance of these tests to other clinical settings. In addition, some studies use comparators that are known to perform poorly, which may exaggerate the performance of certain media.
Performance characteristics of the different media used for screening of rectal or perirectal swabs are shown in Table 6. Of the 13 studies mentioned, 9 showed an almost exclusive presence of KPC producers (80, 139, 141, 150–156), while 2 revealed the exclusive presence of NDM producers (157, 158). Only two studies were done at institutions where KPC and VIM producers were reported to coexist (140, 159), and only one study was performed with OXA-48-producing isolates (160). Furthermore, the prevalence of ESBL producers at these locations is not taken into account and could impact the specificity of these screening methods. The different screening systems performed variably on stool specimens compared with pure cultures (Table 4), showing mostly declines of both sensitivity and specificity with stool specimens.
TABLE 6.
Performance of culture methods on rectal/perirectal swabsa
| Method | No. of positive swabs/total no. of swabs | Sensitivity (%) | Specificity (%) | Enzyme, distribution (%) | Reference standard(s) | Reference |
|---|---|---|---|---|---|---|
| MacConkey agar with IPM | 33/139 | 84.9 | 94.3 | KPC, 100 | PCR for KPC | 80 |
| MacConkey agar with IPM/MEM/ETP disks | 75.8 | 89.6 | ||||
| CHROMagar KPC | 84.9 | 88.7 | ||||
| chromID Carba | 86/177 | 96.5 | 91.2 | KPC, 98; VIM, 1 | qPCR for KPC or VIM, mHT, aminophenyl-boronic acid–meropenem, and EDTA-meropenem testing | 139 |
| MacConkey agar with IPM | 89.5 | 31.9 | ||||
| CDC protocol | 98.8 | 80.2 | ||||
| MacConkey agar with ETP disk | 97/189 | 96.9 | 98.9 | KPC, 61.9; VIM, 9.3; KPC + VIM, 26.8; OXA-48, 3 | Colony PCR for KPC, VIM, OXA-48, IMP, and NDM; negative isolates confirmed with a mHT | 111 |
| MacConkey agar + MEM alone or MEM + PBA, MEM + EDTA, and MEM + PBA | 94.8 | 100 | ||||
| MacConkey agar with MEM | 92/200 | 89.1 | 85.2 | KPC, 68; VIM, 31 | PCR for KPC, IMP, NDM, VIM, and OXA-48; Vitek susceptibility testing and mHT | 140 |
| CDC protocol | 89.1 | 86.4 | ||||
| Enriched brain heart (BH) culture with ertapenem replated onto chromID ESBL agar | 92.4 | 93.3 | ||||
| chromID Carba | 92.4 | 96.9 | ||||
| chromID ESBL | 92.4 | 84.7 | ||||
| chromID Carba | 32/175c | 100 | 98 | NDM, 100 | NDM PCR, disks with meropenem, boronic acid, cloxacillin, and dipicolinic acid (Rosco KPC/MBL Confirm kit) | 157 |
| Brilliance CRE | 59 | 34 | Agar dilution MICs for carbapenems, negative Rosco KPC/MBL test | |||
| CDC protocol | 33/149 | 65.6 | 49.6 | KPC, 100 | KPC PCR; broth microdilution susceptibilities for ertapenem, imipenem, and meropenem; mHT | 150 |
| MacConkey agar with ETP disk with a 27-mm breakpoint | 97 | 90.5 | ||||
| MacConkey agar with IPM, MEM, and ETP disks | 41/122 | 92.7 | 95.9 | KPC, 100 | Direct KPC PCR on swab | 151 |
| CHROMagar KPC | 100 | 98.4 | ||||
| MacConkey agar with MEM and ETP disks | 54/187f | 87 | 100 | KPC, 100 | qPCR on swab and isolates followed by gel electrophoresis and sequencing | 152 |
| MacConkey agar with IPM | 64/755 | 87.5 | 99.4 | KPC, 100 | KPC PCR, KPC PCR, mHT, repeat culture, repeat PCR | 153 |
| chromID Carba (prototype) | 64/37,200c | 100 | 93 | NDM, 100 | PCR for IMP, VIM, GIM, SPM, SIM, and NDM; mHT | 158 |
| Colorex KPC | 97 | 96 | ||||
| CHROMagar KPC | 46/126 | 97.8 | 98.7 | KPC, 72.5; VIM, 27.5 | Phoenix susceptibility; EDTA/IMI confirmatory disks; PCR for KPC and VIM | 159 |
| MacConkey agar with IPM | 78.3 | 97.5 | Negative result not confirmed if negative by both methods | |||
| HardyChrom | 46/126 | 76.1 | 100b | KPC, 100 | qPCRe for KPC and NDM; PCR for SME, VIM, IMP, GES, OXA-48, and AmpC | 141 |
| CDC protocol | 78.3 | 100b | ||||
| Supercarba | 10/77 | 80 | 98.5b | OXA-48, 100 | For positive and negative results, PCRe for KPC, NDM, VIM, IMP, NDM, OXA-48, and ESBL panel | 160 |
| Supercarba with enrichment step | 100 | |||||
| chromID ESBL | 90 | 68.6 | ||||
| chromID ESBL with enrichment step | 100 | |||||
| Brilliance CRE | 80 | 86.6 | ||||
| Brilliance CRE with enrichment step | 100 | |||||
| CHROMagar KPC | 66/95 | 77.3 | 100 | KPC, 100 | Positive results confirmed with PCRe; negative results had negative qPCRe and PCR results | 155 |
| VAN/AMB/CAZ/CLI plate | 77.3 | 100 | ||||
| CHROMagar KPC | 47/150d | 76 | 75.7 | KPC (presumed) | Confirmed with KPC qPCR and Microscan susceptibility testing | 156 |
| SpectraCRE | 97.8 | 86.4 | ||||
| MacConkey agar with ETP disk | 83 | 73.8 | ||||
| CDC protocol | 33/302 | 57.6 | 95.2 | OXA-48, 100 | Initial screen with inhibition disk synergy testing followed by PCR and sequencing of samples with positive results | 165 |
| chromID OXA-48 | 75.8 | 99.3 | ||||
| chromID Carba | 57.6 | 98.9 | ||||
| chromID Carba + chromID OXA-48 | 75.8 | 94.4 |
Abbreviations: MEM, meropenem; IPM, imipenem; ETP, ertapenem; VAN, vancomycin; CLI, clindamycin; CAZ, ceftazidime; AMB, amphotericin B; mHT, modified Hodge test.
Nonfermenting bacteria were excluded.
Stool specimens.
Perirectal swabs.
PCR was done directly on the specimen. If not noted, PCR was performed on pure cultures derived from the sample.
Includes both perianal and perirectal swabs.
We assert here that the sensitivity of a screening medium corresponds to the sum of sensitivities for each particular mechanism (e.g., OXA-48, KPN, and NDM). If a particular medium is tested in cases where one mechanism is overrepresented, it will have a greater contribution to the calculated sensitivity for the detection of CPE. For instance, consider that medium A has sensitivities of 90% for KPC and 70% for OXA-48. If this medium is tested where 95% of CPE are KPC producers while 5% are OXA-48 producers, the study will show an overall sensitivity for CPE detection of 89%. However, if 70% of CPE are OXA-48 producers and 30% are KPC producers, it will show an overall sensitivity of 76%.
To place our analysis in a clinical perspective, we performed a statistical analysis comparing the sensitivities, specificities, and diagnostic odds ratios (DORs) of the different methods used for screening of pure cultures by employing a bivariate random-effects model (161) using the mada package of the R programming language (162, 163). The DOR is the ratio of the odds of the test producing a true-positive result to the odds of it producing a false-positive result. The bivariate random-effects model is a meta-analysis technique for pooling diagnostic performance measures across studies and estimating covariate effects. The corresponding forest plots were generated with ggplot2 (164). Data from methods that did not detect all three carbapenemase classes were excluded. Figures 2 and 3 illustrate the sensitivities, specificities, and DORs for the different media in each study and in aggregate, respectively. Table 5 shows the model-estimated 95% confidence intervals for these parameters. Given the proportion of class B carbapenemase-producing isolates included in these studies, their effect on the estimated pooled performance characteristics is likely disproportionate. The same approach was used to analyze performance on rectal/perirectal swabs (Table 6). Given the low number of specimens for these analyses, we included only data from those studies where KPC was the predominant enzyme (>98%) in the analysis. We excluded methodologies that were not available commercially, except for the CDC protocol. Model-estimated sensitivities, specificities, and DORs with their corresponding 95% confidence intervals are shown in Table 7. Forest plots for the individual and aggregate studies are shown in Fig. 4 and 5.
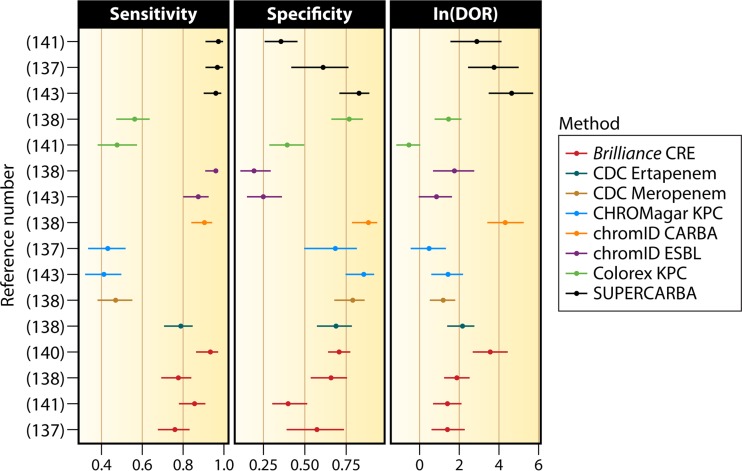
FIG 2.
Per-observation estimates of sensitivity, specificity, and DOR for screening methods used on pure cultures included in statistical analyses (137, 138, 140, 141, 143).
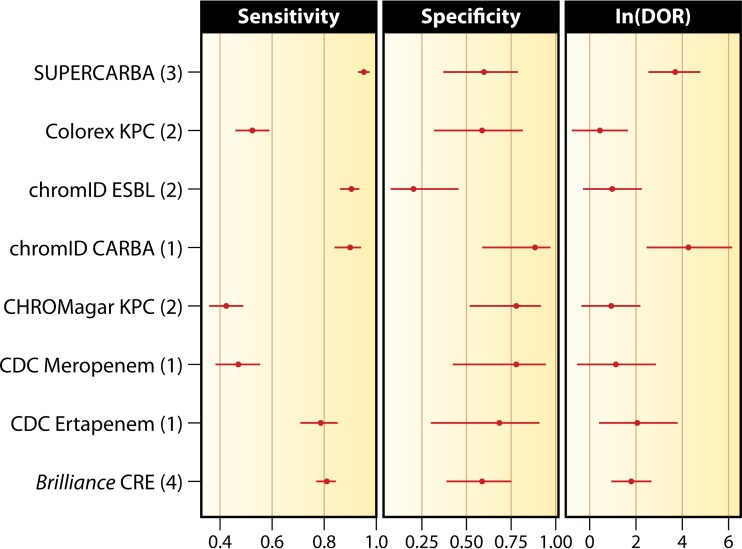
FIG 3.
Aggregate estimates of sensitivity, specificity, and DOR for screening methods used on pure cultures. The number of studies used to calculate the performance of each method is shown in parentheses.
TABLE 5.
Comparison of model estimates of diagnostic performance for different screening methods using pure culturesa
| Method (no. of studies) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | DOR (95% CI) | Aggregate no. of isolates (no. of positive isolates) |
|---|---|---|---|---|
| Brilliance CRE (4) | 81.3 (77.1–84.88) | 58.89 (39.31–76.02) | 6.23 (2.6–14.91) | 774 (439) |
| CDC protocol for ertapenem (1) | 79.23 (71.21–85.47) | 68.57 (30.95–91.4) | 8.32 (1.49–46.57) | 200 (130) |
| CDC protocol for meropenem (1) | 46.92 (38.24–55.8) | 78.57 (42.41–94.81) | 3.24 (0.58–18.19) | 200 (130) |
| CHROMagar KPC (2) | 42.11 (35.69–48.81) | 78.13 (52.18–92.12) | 2.6 (0.73–9.27) | 318 (228) |
| chromID Carba (1) | 90.77 (84.36–94.72) | 88.57 (59.31–97.63) | 76.21 (11.97–485.11) | 200 (130) |
| chromID ESBL (2) | 91.01 (86.26–94.23) | 21.27 (7.95–45.8) | 2.74 (0.75–9.95) | 376 (244) |
| Colorex KPC (2) | 52.45 (45.78–59.04) | 59.13 (32.06–81.6) | 1.6 (0.48–5.35) | 377 (230) |
| Supercarba (3) | 96.27 (93.53–97.87) | 60.38 (37.33–79.59) | 39.29 (12.52–123.33) | 176 (114) |
CI, confidence interval; DOR, diagnostic odds ratio.
TABLE 7.
Comparison of model estimates of diagnostic performance for screening methods for detection of KPC-producing Enterobacteriaceae using rectal/perirectal swabsa
| Method (no. of studies) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | DOR (95% CI) | Aggregate no. of tested swabs (no. of positive swabs) |
|---|---|---|---|---|
| CDC method (3) | 85.37 (58.05–96.09) | 82.94 (41.85–97.05) | 28.37 (0.67–1,209.67) | 452 (165) |
| CHROMagar KPC (4) | 85.16 (61.39–95.4) | 92.74 (70.56–98.55) | 73.35 (1.96–2,743.26) | 506 (187) |
| chromID Carba (1) | 95.98 (65.92–99.66) | 90.76 (30.51–99.55) | 234.36 (4.99–10,996.66) | 177 (86) |
| HardyChrom (1) | 75.53 (22.42–97.06) | 99.38 (72.54–99.99) | 497 (4.9–50,431.85) | 126 (46) |
| SpectraCRE (1) | 96.88 (65.46–99.8) | 86.06 (22.15–99.26) | 191.34 (3.45–10,605.72) | 150 (47) |
CI, confidence interval; DOR, diagnostic odds ratio.
FIG 4.
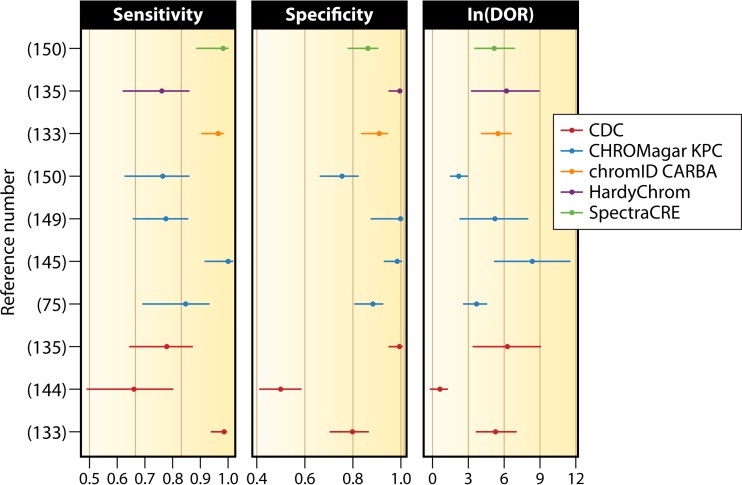
Per-observation estimates of sensitivity, specificity, and DOR for screening of rectal/perirectal swabs (75, 133, 135, 144, 145, 149, 150).
FIG 5.
Aggregate estimates of sensitivity, specificity, and DOR for screening methods using rectal/perirectal swabs. The number of studies used to calculate the performance of each method is shown in parentheses.
Analysis of the results of the screening media in pure cultures shows that chromID ESBL, chromID Carba, and Supercarba have similar sensitivities. The results with Brilliance CRE medium and the CDC method with ertapenem overlap the 95% confidence intervals of chromID Carba and chromID ESBL media. Large confidence intervals can be seen with the CDC method, Colorex KPC, and CHROMagar KPC, reflecting the low number of tested isolates and conflicting results. For instance, CHROMagar KPC performed well in some studies (151, 158, 159) but not all studies (149, 155, 156), with sensitivities ranging from 40 to 98%.
Analysis of specificity is more homogeneous among the different methods. There is, however, a tendency for superiority favoring chromID Carba, while the opposite holds for chromID ESBL. This is expected, as the growth of ESBL-producing Enterobacteriaceae is considered a false-positive result when screening for CPE. Supercarba had a wide range of specificities, ranging from 35% to 82% depending on the details of the analysis, which is reflected in its large confidence interval. In this analysis, chromID Carba and Supercarba have a clear advantage in the clinic compared to the other methods. Given the large confidence intervals, these results must be interpreted with caution. Not included in the above-described analysis are data from a study by Hu et al., as it involved only 18 isolates of KPC-producing Enterobacteriaceae (105).
Analyses of medium performance on rectal/perirectal swabs are limited to those studies where KPC was the prevailing enzyme. Unfortunately, there are not enough data for a meaningful comparison of these media under different conditions. Available data with note of the enzyme distribution can be found in Table 6. The sensitivities for detection of KPC in rectal/perirectal swabs show overlapping confidence intervals for all methods, except for the CDC protocol, which is clearly inferior. Specificities also show significant overlap. HardyChrom agar showed the worst specificity, although it had a very large confidence interval, a product of being tested in only one study (141). MacConkey agar with imipenem also performed acceptably in some studies (80, 111, 139, 150), showing sensitivities and specificities as high as 92% and 100%, respectively. Analysis of DORs shows homogeneity for most methods. The overall trend is for the CDC method to be inferior to the others. Improved performance is suggested for SpectraCRE, HardyChrom, and chromID Carba. The confidence intervals for HardyChrom and SpectraCRE, however, are exceedingly large. SpectraCRE was tested in a single study in a Chicago LTCF (156), which likely explains its large confidence interval.
Due to its limited scope, chromID OXA-48 agar was not included in this statistical analysis. In a study by Zarakolu et al., it showed 75% sensitivity when tested against clinical specimens containing OXA-48, with 99.3% specificity. When used in conjunction with chromID Carba, the sensitivity and specificity reached 90.9% and 98.5%, respectively (165).
The overall differences in sensitivity between the media can be explained by the carbapenemase being tested. Most media perform reasonably well with class A enzymes, while performance with class B and D enzymes is more variable. chromID Carba medium performed well both in pure culture and when tested on rectal/perirectal swabs. The Supercarba medium performed well on pure cultures. However, it was not tested with patient specimens. SpectraCRE performed well on rectal/perirectal swabs, although one must be aware of its confidence interval. The CDC method underperformed when tested on pure cultures and clinical specimens. Other methods that were tested, particularly those involving “house-grown” techniques, could not be analyzed with the same rigor, and unless more studies are done, we caution against their use in clinical practice.
It must be emphasized that many of the studies of these selective plates are limited to KPC-producing isolates. Furthermore, the various media evaluated in Tables 4, 6, and 8 are not available in all countries. Therefore, the practical issues of cost and availability affect the choice made by an individual laboratory, which must decide if optimal sensitivity is desired, knowing that additional workup will be required to detect false-positive results if the method has a low specificity.
TABLE 8.
Geometric mean limits of detection in pure cultures according to culture method and β-lactamase classa
| Method | Geometric mean limit of detection (CFU/ml) |
No. of isolates of class A/B/D | Reference | ||
|---|---|---|---|---|---|
| Class A | Class B | Class D | |||
| Supercarba | 1.47 × 101 | 3.36 × 101 | 1.37 × 101 | 18/52/44 | 149 |
| chromID ESBL | 1 × 101 | 1.26 × 101 | 1.21 × 103 | 18/52/44 | |
| chromID KPC | 8.1 × 101 | 1.64 × 103 | 3.43 × 106 | 18/52/44 | |
| Supercarba | 1.41 × 101 | 2.81 × 101 | 1.59 × 101 | 20/51/43 | 143 |
| Brilliance CRE | 1.12 × 102 | 2.86 × 102 | 1.45 × 103 | 20/51/43 | |
| CHROMagar KPC | 5.89 × 101 | 8.93 × 102 | 4.62 × 106 | 20/51/43 | |
| MacConkey agar with imipenem | 4.68 × 102 | 1.24 × 103 | NT | 8/2/0 | 80 |
| MacConkey agar with meropenem/ertapenem disks | 2.62 × 106 | 3.32 × 105 | NT | 8/2/0 | |
| CHROMagar KPC | 2.02 × 103 | 1.24 × 104 | NT | 8/2/0 | |
| CDC method | 6.87 × 101 | 8.66 × 102 | 5.2 × 107 | 5/2/1 | 140 |
| chromID ESBL with prior enrichment in BHI broth + 10 μg ertapenem | 2.6 × 101 | 5.55 × 101 | ND | 5/2/1 | |
| chromID ESBL | 7.49 × 101 | 4.42 × 102 | ND | 5/2/1 | |
| chromID Carba | 2.11 × 101 | 4.42 × 102 | 5.5 × 107 | 5/2/1 | |
| Brilliance CRE | 2.67 × 101 | 3.41 × 101 | 3.77 × 101 | 12/14/5 | 166 |
| chromID OXA-48 | 1 × 107 | ND | 3.36 × 101 | 10/10/57 | 167 |
| chromID Carba | 1 × 101 | 2.0 × 101 | 1.62 × 104 | 10/10/57 | |
| Supercarba | 3.16 × 101 | 2.51 × 102 | 2.98 × 101 | 10/10/57 | |
| CHROMagar KPC | ND | ND | 1.26 × 104 | 0/0/9 | 100 |
| CHROMagar ESBL | ND | ND | 5.26 × 103 | 0/0/9 | |
NT, not tested; ND, not detected; BHI, brain heart infusion.
Studies analyzing the LOD include bacteria with specific genetic backgrounds on pure cultures that may not necessarily represent the backgrounds present in a specific clinical setting (Table 8). The LOD will directly impact the sensitivity of the screening method. Given the abundance of Enterobacteriaceae in stool, it is desirable to inhibit the growth of the carbapenem-susceptible population. However, this inhibition comes at the expense of sensitivity. A relatively low inoculum of a CPE isolate with borderline susceptibility will need to overcome this inhibitor, and the medium would have a higher LOD. On the other hand, adjusting growth inhibitors to obtain a lower LOD would allow for the growth of other bacteria and would decrease specificity.
High-resource settings where health care is already expensive may have a lesser impact on the isolation of more patients and may want to err on the side of higher sensitivities. Furthermore, the medical care provided in high-resource settings tends to be more invasive; therefore, there is a higher cost of missing a colonized patient. On the other hand, lower-resource settings may still benefit from the selection of a method with lower sensitivity that would decrease isolation costs while still having an impact on the local spread of CPOs.
Table 8 summarizes the limits of detection of the different agar screening media. All of the tested media and Supercarba agar performed reasonably well for the detection of class A enzymes (KPC), achieving a LOD in the range of 1 × 101 to 1 × 102 CFU/ml (80, 140, 143, 149, 166), except for chromID OXA-48, which, as expected, performs better with class D enzymes (OXA-48) (167). The LOD for class B enzymes on chromogenic media are ∼1 log higher than those for class A enzymes (80, 140, 143, 149). MacConkey agar with disks had a LOD ∼1 to 2 logs higher than those of the other media for both class A and B enzymes (80, 140). chromID OXA-48 showed poor performance for both class A and B enzymes, with a LOD of 1 × 107 CFU/ml (167). Class D enzymes were not tested by all methods. The LOD for Supercarba remained consistently close to 1 × 101 CFU/ml. Other methods showed a significant increase in their LOD for class D enzymes. Remarkably, chromID KPC, CHROMagar KPC, the CDC method, and chromID Carba had a LOD up to 6 logs higher than those of more sensitive methods (140, 143, 149). As expected, chromID OXA-48 performed exceptionally well with class D enzymes, with a LOD of 5 × 101 CFU/ml (167).
Nucleic acid amplification technology.
Nucleic acid amplification technology (NAAT) detects the presence of a specific gene or genes, in most cases limiting its usefulness to previously characterized determinants. Furthermore, newly emergent variants of previously characterized genes may not be reliably detected. Since various genes can encode different carbapenemases, a broad panel of tests is needed to detect all targets. Because it is not practical to detect every enzyme, these tests have been designed to cover the most common carbapenemases. A challenge for nucleic acid-based testing is DNA extraction from stool. Feces contain PCR-inhibiting substances, and poor results may be obtained due to excessive shearing of DNA (168). Despite these concerns, very good methods are available for extracting DNA from stool, and multiplex molecular assays are routinely performed on stool specimens for gastrointestinal pathogens. It is critical to note that detection of resistant determinants in pure cultures or in specimens where a single organism is expected (such as blood or urine) is significantly easier than detection of the same genes in a more complex specimen such as a stool swab. In addition, epidemiological data such as species information are lost in most assays.
There are several NAAT-based methodologies that may be employed to detect carbapenemase genes in bacterial isolates (169). Theoretically, all of them can be used to screen stool specimens. These methodologies include single- and multiplex endpoint PCRs, loop-mediated isothermal amplification (LAMP), single- and multiplex quantitative real-time PCR (qPCR), and microarrays. Next-generation sequencing (NGS) may be another option, although it is not readily available in most clinical laboratories at this time (169). NGS remains expensive due to high equipment acquisition costs and the need for significant computer processing power and data storage (170).
Regardless of the NAAT-based methodology selected by a laboratory, there are complex regulatory requirements that vary from region to region. Implementation of a laboratory-developed assay involves the determination of the test's performance characteristics. The burden of an involved development-and-validation process may be partially relieved by the use of commercial assays. U.S. Food and Drug Administration (FDA) regulations in the United States are evolving at this time and will likely result in an increased regulatory burden on laboratories in the future.
Endpoint PCR is useful when there is a large quantity of the target gene. Specificity cannot be ensured unless positive results are confirmed by DNA sequencing or hybridization with specific probes. With proper validation, a PCR method can be acceptable. The Hyplex Super Bug ID system (Amplex Biosystems GmbH, Giessen, Germany) for the detection of carbapenemases is based on a multiplex endpoint PCR followed by enzyme-linked immunosorbent assay (ELISA) hybridization (171). Although it has not been tested directly on stool specimens, this NAAT showed 98% sensitivity and specificity for VIM-producing CPE when used on DNA extracted from clinical specimens, including blood, urine, pus, and respiratory samples, from Greece (172). Another multiplex endpoint PCR was developed by Voets et al. and allows the detection of a wide range of resistance genes (173). Some of these multiplex assays were developed by independent laboratories and are not widely available to most clinical laboratories. However, there is great value in demonstrating that these comprehensive assays can be developed.
Microarrays consist of oligonucleotides bound to a solid surface. The target gene of the pathogen is then labeled and hybridized to the immobilized probe. This reaction is then measured with a scanner (169). Microarrays are difficult to standardize (169), and reports describing the use of microarrays to directly screen for β-lactamase genes in stool are not available. Most assays, however, can be used to confirm and characterize the β-lactamase gene from suspicious colonies of a screening culture. These tests have excellent sensitivity and specificity, as shown by a study with 149 previously characterized Enterobacteriaceae that were subjected to a commercial Check-Points microarray assay (Check-Points Health, Wageningen, Netherlands), which was found to have 100% sensitivity and specificity (174). Direct testing of blood cultures also showed 98% concordance between a microarray method and routine microbiological testing (175). The Verigene BC-GN test is a microarray-like detection system. It detects nine genus/species targets and six resistance determinants, including KPC, NDM, OXA, VIM, and IMP, without the need for prior PCR amplification (176). Future studies are needed to determine if microarrays will be used to screen perirectal or stool specimens directly, although this may be hampered by their high cost and the advent of next-generation sequencing.
Loop-mediated isothermal amplification (LAMP) is a modification of conventional PCR where several oligonucleotides that bind to the target gene are incubated at the same temperature with the DNA polymerase. As DNA polymerizes, there is a release of pyrophosphate that can be detected with a fluorescent dye or a compound that will increase the turbidity of the solution (177). This method's advantages include increased sensitivity with low DNA concentrations compared to endpoint PCR, no need for a thermocycler, and simple visualization of the result. LAMP assays can be particularly useful in low-resource settings (169). A LAMP assay for the detection of NDM-1 was successfully used on 336 clinical specimens, including rectal swabs (178). Those investigators found a limit of detection of 10.70 pg/μl of genomic DNA, which would correspond to roughly 1 × 103 CFU, compared to 1,070 pg/μl (or 1 × 105 CFU) for the endpoint PCR assay used as a comparator in that study. Solanki et al. developed two LAMP assays for the detection of KPC and NDM-1 (179). These assays were able to detect all 48 tested isolates with either NDM or KPC, while endpoint PCR detected only 44. Other studies have found an improved performance of LAMP versus endpoint PCR for microbiological targets other than CPE but not versus real-time assays (180, 181). Therefore, LAMP assays may have a useful role in detecting CPOs, but they are not the most sensitive assay for clinical microbiology laboratories that have access to other types of NAATs.
Real-time or quantitative PCR (qPCR) is based on the coupling of PCR with the detection of the amplified target. Real-time PCR has been used for the screening of CPO using both commercial and “in-house” kits, with the advantage of more rapid results, increased sensitivity, and increased specificity (152, 153, 182, 183). A recent seven-center study in the Netherlands found 100% sensitivity and specificity for a multiplex assay used for the detection of KPC, NDM, VIM, IMP, and OXA-48 in 20 selected laboratory isolates (184).
Many laboratories have experience in using qPCR for the direct screening of stool/rectal swab specimens. Examples of stool/rectal swab testing with qPCR in routine clinical practice include screening for vancomycin-resistant enterococci, group B streptococci, and Clostridium difficile (185–187). The validity of the use of qPCR, including for quantification of KPC carriage loads, was evaluated by Lerner et al. (188). They determined a detection limit of 10 plasmid copies, which we presume is close to 1 × 101 CFU/ml.
Another in-house qPCR for NDM-1 found a limit of detection of 1 × 101 to 3 × 101 CFU/ml of stool, compared to 2 × 101 to 1 × 102 CFU/ml for chromID ESBL and 2 × 101 to 4 × 103 CFU/ml for CHROMagar KPC (189). An additional study found limits of detection with endpoint PCR of 1 × 104 to 1 × 105 CFU/ml for KPC and 1 × 103 CFU/ml for NDM (141). Using an in-house qPCR assay, Naas et al. found a limit of detection for OXA-48 of 1 × 101 to 1 × 102 CFU/ml in stool for qPCR, compared to 1 × 101 to 1 × 102 CFU/ml for Supercarba and 2 × 101 to 3 × 102 CFU/ml for chromID ESBL (182). A comparison between agar screening and qPCR for KPC showed 100% sensitivity for the qPCR assay, compared to 77% for culture methods (155). Overall, the limits of detection for single-gene assays (152, 155, 182, 190) tend to be lower than those for multiplex assays (141, 191).
Commercial assays for molecular multiplex CPO detection include Check-Direct CPE, Check-MDR Real Time (Check-Points Health), Hyplex SuperBug ID (Amplex Biosystems), Eazyplex SuperBug CRE (Amplex Biosystems), and Xpert MDRO (Cepheid). Check-MDR Real Time consists of an oligonucleotide probe that binds to the target sequence (VIM, NDM, KPC, and OXA-48), to a pair of universal primers, and to a molecular beacon. Real-time PCR amplifies only the bound target sequences at the same time that the molecular beacon emits fluorescence to measure amplification. The manufacturer has established a limit of detection of <5 copies per reaction. Testing of pure cultures showed 100% sensitivity and specificity (192). The manufacturer, however, recommends the use of pure cultures, which is clearly not the method used to screen perirectal swabs directly. Check-Direct CPE is a real-time assay using probe detection chemistry. It has a limit of detection of 5 copies per reaction. Using “spiked” stool specimens, Check-Direct CPE was able to detect a bacterial inoculum of 103 to 105 CFU/ml, with less sensitivity for KPC (191). NucliSENS EasyQ KPC (bioMérieux) is another real-time assay that uses molecular probes. This assay was compared to chromID ESBL with ertapenem disks by using surveillance specimens. Although a limit of detection was not determined, the assay showed 93% sensitivity (193). The SuperBug CRE system is a multiplex LAMP system that is able to detect KPC, VIM, NDM, and OXA-48 (and some variants) in addition to the ESBLs CTX-M-1 and CTX-M-9. On pure cultures, Eazyplex SuperBug CRE was able to correctly identify all 139 Enterobacteriaceae isolates (194). However, when used against a panel of 82 Acinetobacter species isolates, it produced 5 false-positive results (195). The Xpert MDRO assay has been used to detect CPO directly from rectal and perirectal swabs. The assay was able to detect KPC, NDM, and VIM with 100% sensitivity and 99% specificity using 328 discarded perirectal, rectal, or stool samples from two U.S. hospitals and one Spanish hospital (183).
Table 9 summarizes the molecular methods that have been used on clinical specimens. Some of the reports show excellent sensitivity and specificity for molecular assays; however, other studies show a broad range for the LOD of specific carbapenemases, with numbers comparable to those for agar-based methods. Limitations include the cost and the inability to detect new or unanticipated carbapenemases. Pooling of specimens for initial screening, followed by confirmatory testing of positive samples, may be an option to contain costs, especially in low-prevalence settings, but investigations are needed to see if the loss of sensitivity is too great. The cost-effectiveness of the approach will also depend on the prevalence of CPOs at an individual institution, which will determine the number of specimens that would require follow-up testing when a positive pooled result is obtained, because all individual specimens in the pool would need to be retested. The concern about detecting new, not-yet-described, carbapenemases will need to be addressed by constant vigilance in updating targets in a chosen assay; if a laboratory reports a new carbapenemase in the local geographic region, or when a medical center treats a large volume of international patients, adjustments will need to be made.
TABLE 9.
PCR-based testing for CPOs in clinical specimens
| Target(s) | Methodology | Specimen type(s)b | Sensitivity (%) | Specificity (%) | Limit(s) of detection (CFU/ml) | No. of specimens | Reference |
|---|---|---|---|---|---|---|---|
| KPC | PCR | ES | 92.2 | 99.6 | Not calculated | 755 | 153 |
| KPC | qPCR | ES | 97 | 96.6 | 4 × 100–4 × 102 | 95 | 155 |
| KPC | qPCR | S, MC | 100 | 98 | 5 × 100 CFU/ml | 216 | 190 |
| VIM, IMP, KPC, OXA-48, NDM-1 | PCR (Hyplex SuperBug ID) | MC | 98.0 | 98.6 | Not calculated | 236 | 172 |
| OXA-48 | qPCR | PC, SS | 100 | 100 | 101–102 | 35 | 182 |
| KPC, NDM-1 | qPCR | S, SS | 100 | 100 | KPC, 104–105; NDM, 103 | 46/80a | 141 |
| KPC | qPCR (EasyQ KPC) | S | 93.3 | 99 | Not calculated | 806 | 193 |
| VIM, OXA-48, NDM, KPC | qPCR (Check-MDR Carba) | SS | 100 | 100 | 103–105 | 25 | 191 |
| KPC, NDM, VIM | qPCR (Xpert MDRO) | S, SS | 100 | 99 | <3 × 102 | 328 | 183 |
| KPC | qPCR | S | 97.9–100 | 95–96.4 | 5 × 100 | 187 | 152 |
| NDM-1 | qPCR | SS | 100 | Not calculated | 101–3 × 101 | 32 | 189 |
Specificity panel of 80 known negative specimens.
ES, stool or perirectal swab with prior enrichment; S: stool, stool swabs, or perirectal swabs; MC, mixed clinical specimens; SS, spiked stool or stool swabs; PC, pure cultures.
SCREENING OPTIONS
As discussed above, there are several screening options that may be easy to implement in a clinical microbiology laboratory. Any of these options should be closely coordinated with the infection control program of the institution. One must know the baseline prevalence and type of resistance enzymes in a specific setting, as the choice of method will be dependent on these variables. At this time, we cannot find data to suggest any advantage of using stool specimens versus rectal or perianal swabs. Most studies have been done using rectal swabs, and it is likely that most institutions would tend to prefer this modality.
Whatever modality is chosen for screening, laboratories must be aware that local validation will be required. When implementing a screening program, it is important to consider certain factors (Table 10). The decision regarding who should be screened will always be controversial, as the balance of cost and risk will be subject to different interpretations. Ideally, screening should be universal, but most institutions do not have the necessary resources for both screening and isolation while waiting for test results. Table 11 proposes a set of criteria that can be used to screen certain patient populations.
TABLE 10.
Factors to consider when implementing a screening program
| Factor |
|---|
| Epidemiology of CPO in the community |
| Prevalence of each carbapenemase |
| Ability to identify high-risk groups |
| Availability and cost of isolation beds |
| Existing logistics for collecting specimens (e.g., an existing screening program for carriers of vancomycin-resistant Enterococcus spp.) |
| Current clinical microbiology laboratory capabilities |
| Availability of molecular diagnostic tools |
| Available technologists and ability to accommodate testing |
| Experience developing and validating in-house assays |
| Experience/availability of other technologies to detect carbapenemases |
| Carba NP or Blue-Carba |
| Inhibition disk assays (double-disk synergy test) |
| UV spectrometry or MALDI-TOF MS |
| Modified Hodge test |
TABLE 11.
Proposed criteria for screening of patients for CPO infection
| Area | Patients for screening |
|---|---|
| Areas where the disease is not endemic | Patients with multiple hospital admissions |
| ICU patients | |
| Patients who have received medical care in areas of endemicity over the last 12 months | |
| Patients who reside in health care settings | |
| Patients with history of CPO infections or colonization | |
| Patients with prior prolonged hospital stays | |
| Patients coming from areas of endemicity | |
| Patients who are or who are expected to become incontinent or unable to take care of their personal hygiene | |
| Areas where the disease is endemic | Everyone (as resources for testing, isolation, and cohorting allow), with particular emphasis on critically ill patients, patients who are unable to take care of their excreta, and patients with an expected prolonged hospital stay |
Culture-Based Screening with Molecular Confirmation
Culture-based screening includes the use of a chromogenic agar, Supercarba, or the CDC method to perform perirectal or rectal swab screening of patients. While universal screening has a higher potential for detection and prevention of outbreaks, it comes at a significant financial cost. Based on our analysis, we favor the use of chromID Carba. However, if the hospital is located in a geographic area with a high incidence of OXA-48, the clinical microbiology laboratory should strongly consider the use of Supercarba or the addition of an OXA-48-specific medium such as chromID OXA-48 medium. A biplate containing chromID Carba and chromID OXA-48 is available (chromID Carba Smart; bioMérieux, France). Bacteria that grow on Supercarba medium should be identified by conventional microbiological tests or MALDI-TOF MS. Similarly, isolates on chromogenic media that cannot be readily classified as Enterobacteriaceae should also be identified. While non-Enterobacteriaceae can carry plasmids encoding carbapenemases, commonly, carbapenem resistance in these organisms is mediated by other mechanisms. The decision to isolate patients with carbapenem-resistant organisms other than Enterobacteriaceae should be based on local epidemiology. Confirmation of positive specimens should ideally be sought with molecular testing with either a broad panel of PCRs or a microarray method. Alternatively, a phenotypic test (such as Carba NP, Blue-Carba, inhibitory disk synergy testing, or mHT) can be used to confirm the presence of carbapenemases, reserving molecular testing for a random sample of positive isolates. Random sampling will come at the cost of decreased hospital epidemiology data. To track the prevalence and type of carbapenemases in an institution, isolates from clinical specimens, not only surveillance specimens, demonstrating decreased carbapenem susceptibility should also be subjected to an assay for the detection of carbapenemase activity or a PCR panel/microarray.
Figure 6 proposes an algorithm for the use of a culture-based approach. Note that depending on the laboratory capabilities, some samples may be referred to a research laboratory.
FIG 6.
Screening by conventional microbiology.
Molecular-Based Approach
The use of universal screening of perirectal swab samples via molecular methods may not be desirable or affordable due to low prevalence or increased costs. Screening of high-risk patients, such as those returning from areas of endemicity, those who have been transferred from LTCFs, or those who have had extensive exposure to carbapenems (41), may be advisable. A multiplex real-time PCR assay that includes KPC, OXA-48, NDM, and VIM should be used in most locales. However, laboratories in specific areas where IMP or GES-5 is common should either develop their own assays or perform simultaneous routine culture testing. Specimens with indeterminate results and a sample of negative specimens obtained through molecular testing should be tested with a culture-based method with high sensitivity, such as Supercarba or chromID Carba. Suspect colonies should be subjected to antimicrobial susceptibility testing or to a test for carbapenemase activity (e.g., Carba NP or Blue-Carba). A test such as the double-disk synergy test with avibactam-ertapenem with follow-up testing with ertapenem-boronate or moxalactam-EDTA, depending on the result, may enable a laboratory to distinguish between class A, B, and D enzymes (113), making it particularly attractive for this scenario. Figure 7 suggests an algorithm for CPO screening based on molecular methods.
FIG 7.
Molecular screening algorithm.
Combined Approaches
Some combined approaches can be useful in specific situations, such as during an outbreak caused by a CPO with a known enzyme. We speculate that a LAMP or qPCR assay could be implemented for universal screening of carbapenemases involved in an outbreak. Patients who test positive could be quickly cohorted, while patients who test negative could be subjected to routine culture-based screening. This strategy would maximize the number of available hospital beds while attempting to minimize the number of patients under enhanced infection control precautions.
CONCLUSIONS
In this review, we stress that screening for intestinal carriage of CPOs is of significant importance for the development of infection control strategies. However, the optimal screening modality remains to be established for each location and for each specific purpose. Culture-based screening methods have the advantage that they involve technologies that are readily available in clinical microbiology laboratories. Some enhancements, such as the use of chromogenic media, make culture-based screening more convenient; however, the turnaround time is long, and the sensitivity of some culture methods is not as high as desired. In addition, culture-based methods may not be optimal for the detection of low-level carbapenemase production, which is important for epidemiological purposes (93).
Agar-based procedures always require confirmatory testing to detect the type of bla gene present after a potentially resistant isolate is detected. Clinical microbiology laboratories may choose an agar-based screen with follow-up molecular testing or a molecular method with reflex to culture if further investigation of the isolate is desired. On the other hand, NAATs offer a promising approach for screening for carriage of CPOs. These methods offer faster availability of results and increased sensitivity but with a significantly increased expense and an unclear specificity with direct specimens at this time.
Our review indicates that chromID Carba and Supercarba media have excellent sensitivities for class A β-lactamases that rival that of real-time PCR. As there are more communities where KPC is becoming endemic, the use of screening for this class of enzymes may become less useful because the high prevalence could make empirical therapy and initial isolation procedures prior to surveillance results default to the assumption of a KPC-positive isolate. It is still hoped, however, that communities in which KPC-positive organisms are not endemic may contain the spread of resistant isolates for some time. We argue that screening should shift to those carbapenemases that are threatening to become common and that have a high potential of causing outbreaks, such as NDM and OXA-48. Real-time PCR appears to be ideally suited for this goal; however, qPCR implementation is hampered by cost. In addition, there could be false-positive results for OXA-48 due to the amplification of similar chromosomally encoded enzymes in species such as A. baumannii. Furthermore, data on their performance in the clinical setting compared to that of culture-based screening are not yet available. Nonetheless, the improved turnaround time and improved accuracy of NAATs and direct carbapenemase detection assays may result in limiting the unnecessary prolonged isolation of newly admitted patients, thus decreasing costs to the infection control program. Nevertheless, molecular tests with high sensitivity have a cost that is difficult to offset and can be prohibitive for many clinical microbiology laboratories (136). We must choose wisely.
There is an urgent need to define the appropriate criteria and clinical circumstances to conduct screening for gastrointestinal carriage of CPOs and to determine the optimal methods to be used. The best method will differ from institution to institution depending upon the prevalence in the community, the travel patterns and demographics of the population, the level of care rendered by the hospital, the age of patients, the technical capabilities of the microbiology laboratory, and the resources allocated by the hospital administration for infection control monitoring.
Agar-based and colorimetric screens are usually more affordable and are able to detect the presence of “new and emerging” carbapenemases before they are characterized. In addition, most agar-based tests can be performed without the need for significant investments by the clinical microbiology laboratory. However, they may lack sensitivity for carbapenemase producers that confer a low level of resistance. Furthermore, variability in performance according to the β-lactamase class makes the selection of a particular method more difficult. For instance, it is easier to justify the resources needed to perform a screening test that will reliably detect class A carbapenemases in an area where KPC is endemic. At the same time, the detection of class D carbapenemase producers may allow for the institution of a program that will prevent them from becoming prevalent. Laboratories may consider a combined approach of two independent assays to screen for a broader spectrum of carbapenemases. This strategy, however, comes with the disadvantage of increased cost and labor.
At this time, clinical microbiology laboratories that choose to implement a CPO screening methodology must have a reliable procedure for the detection of the carbapenemases in their area of endemicity. In most of the United States, this carbapenemase would be KPC, while laboratories in Europe and the Middle East should likely screen for OXA-48. Once a bla gene is found, laboratories should choose which of the other carbapenemases that they want to include in their screening approach, knowing that they will miss colonized patients carrying a CPO not included in their infection control algorithm.
Although this review focuses on carbapenemase-producing Enterobacteriaceae, plasmids carrying carbapenemase genes have been identified in non-Enterobacteriaceae, so vigilant monitoring of both types of organisms may be warranted in the future. We assert that until a carbapenem-resistant isolate is recovered in a clinical culture and detected by routine susceptibility testing, the number of carbapenemase targets to be included will be determined by cost, time available for labor, and technical abilities of the laboratory. It is likely easier to adapt an in-house molecular assay than a commercial assay to detect emerging carbapenemases by adding additional primers and probes to an existing assay, which provides information about both the presence of enzymes and the type of enzyme. In contrast, commercially available assays have the advantage of manufacturer validation, but they cannot be modified quickly and are modified only when the manufacturer chooses to update the assay. Continued surveillance to detect carbapenemases not detected with the chosen assay is warranted. These considerations are most important to prevent and control infections caused by carbapenemase producers and protect public health. A proactive approach to halt the spread of carbapenemase producers is desperately needed.
ACKNOWLEDGMENTS
Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award numbers R01AI100560, R01AI063517, and R01AI072219 to R.A.B.; NIAID DMID protocol 10-0065 to K.K.; the Intramural Research Program of the NIH, the Cleveland Department of Veterans Affairs, Veterans Affairs Merit Review Program Award 1I01BX001974, and Geriatric Research Education and Clinical Center VISN 10 to R.A.B.; and the Swiss National Science Foundation under grant number 32003B_153377 to A.E.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biographies

Roberto Viau was born in Guatemala City, Guatemala. He obtained his medical degree from the Universidad Francisco Marroquin, where he graduated cum laude in 2008. He has been involved in Gram-negative resistance since then. Dr. Viau completed an Internal Medicine Residency at the Jacobi Medical Center in New York, NY, and is currently an Infectious Diseases Fellow at Case Western Reserve University. He is interested in molecular diagnosis and the epidemiology of resistance mechanisms. He will be joining the faculty at Case Western Reserve University upon completion of his fellowship. He will continue working on Gram-negative resistance as well as on diagnostic microbiology.

Karen M. Frank is the Chief of Microbiology for the National Institutes of Health Clinical Center. She completed her M.D. and Ph.D. in Biochemistry at the University of Pennsylvania in 1994, studying magainins and discovering the antimicrobial molecule squalamine. She completed a Clinical Pathology Residency at the Brigham & Women's Hospital and conducted postdoctoral research on V(D)J recombination in a Howard Hughes Medical Institute laboratory at Boston Children's Hospital. She was a Pathology faculty member at the University of Chicago for 12 years before relocating to the NIH in 2012, investigating DNA repair pathways as well as staphylococcal toxins. In collaboration with investigators at the National Human Genome Research Institute and the Clinical Center, she is using DNA sequencing to characterize carbapenem-resistant Enterobacteriaceae, tracking infections and the spread of resistance genes on plasmids between bacteria, as well as conducting in vitro analyses of the bacterial conjugation of several outbreak strains.

Michael R. Jacobs, M.D., Ph.D., received his medical degree and Medical Microbiology doctorate from the University of the Witwatersrand in Johannesburg, South Africa, where he also did his Medical Microbiology Residency. He is a diplomate of the American Board of Medical Microbiology, Member of the Royal College of Pathologists, Professor of Pathology and Medicine at Case Western Reserve University, and Director of Clinical Microbiology at the University Hospitals Case Medical Center. Dr. Jacobs' research interests include epidemiology, antimicrobial susceptibility, and mechanisms of resistance of antibiotic-resistant Streptococcus pneumoniae and carbapenem-resistant Enterobacteriaceae and non-Enterobacteriaceae. He has served as an Examiner for the American Board of Medical Microbiology and as an Editorial Board member of Antimicrobial Agents and Chemotherapy and is currently on the Editorial Board of the Journal of Clinical Microbiology. Dr. Jacobs has presented numerous abstracts at national and international conferences as well as authored over 400 papers in peer-reviewed journals.

Brigid Wilson has a B.A. in Mathematics from Swarthmore College and a Ph.D. in Statistics from UCLA. She currently works at the Louis Stokes VA Medical Center in Cleveland, OH, under the Geriatric Research, Education and Clinical Center and provides statistical support to VA-affiliated researchers. She has previously provided statistical support to researchers while working for UCLA's Statistical Computing Group and to pharmaceutical, biodefense, and biomedical companies while working in private biostatistics consulting.

Keith Kaye is a Professor of Medicine in the Division of Infectious Diseases and Department of Medicine at Wayne State University and Detroit Medical Center (DMC). He is Corporate Vice President of Quality and Patient Safety and the Corporate Medical Director of Hospital Epidemiology and Antimicrobial Stewardship at DMC. Dr. Kaye's particular academic interests and skills are in the epidemiology of and outcomes associated with multidrug-resistant bacteria, infections in the elderly, surgical-site infections, device-related infections, and antimicrobial stewardship. Dr. Kaye received his medical degree from the University of Pennsylvania and served his Internal Medicine residency and was an Infectious Diseases fellow at Beth Israel Deaconess Medical Center in Boston, MA. During his fellowship, Dr. Kaye earned a master's degree in Public Health from the Harvard School of Public Health. Dr. Kaye is currently the principal investigator of a multicenter NIH-funded contract studying polymyxin-based therapy for infections due to extremely drug-resistant Gram-negative bacilli.

Curtis J. Donskey received his M.D. from the Medical College of Wisconsin in 1990. He completed an Internal Medicine Residency and Chief Residency at Brown University and then completed an Infectious Diseases Fellowship at the University Hospitals of Cleveland. He is currently the Chairman of the Infection Control Committee at the Cleveland Veterans Affairs Medical Center and Associate Professor of Medicine at Case Western Reserve University. His research focuses on infection control and the role of intestinal colonization in the spread of resistant bacteria in hospital settings.

Federico Perez is an Assistant Professor of Medicine at Case Western Reserve University School of Medicine and a member of the medical and research services at the Louis Stokes Cleveland Department of Veterans Affairs Medical Center. He is currently a scholar at the Clinical and Translational Science Collaborative of Cleveland. He first became interested in the molecular epidemiology and transmission dynamics of multidrug-resistant Gram-negative bacteria in 2002 while working at the Centro Internacional de Entrenamiento e Investigaciones Medicas in Cali, Colombia.

Andrea Endimiani obtained a medical degree (1999), Board in Medical Microbiology and Virology (2003), and a Ph.D. in Immunopathology (2007) from the University of Insubria (Varese, Italy). Dr. Endimiani has been trained in molecular microbiology at the Antibiotic Management Program of the University of Pittsburgh (2006 to 2007) and the Case Western Reserve University School of Medicine in Cleveland (2007 to 2010). Currently, he is a Researcher and Medical Microbiologist at the Institute for Infectious Diseases of the University of Bern (Switzerland). Dr. Endimiani's group focuses on the genetic background of antibiotic-resistant Gram-negative pathogens (particularly those producing ESBLs and/or carbapenemases); this group is also very interested in the development of new strategies (e.g., microarrays and multiplex real-time PCRs) to speed up the detection of infections due to multidrug-resistant bacteria and sexually transmitted diseases (mainly those due to Neisseria gonorrhoeae). Dr. Endimiani was recently (2014) qualified as a full Professor of Microbiology and Medical Microbiology.

Robert A. Bonomo received his M.D. from the Case Western Reserve University School of Medicine. He trained in Internal Medicine and Infectious Diseases at the University Hospitals of Cleveland and received a Certificate of Added Qualification in Geriatrics. Dr. Bonomo served as section chief of the infectious disease division at the Cleveland Veterans Affairs Medical Center before becoming Director of the Veterans Integrated Service Network 10 Geriatric Research, Education and Clinical Center (GRECC). He also serves as Chief of Medicine. He is a Professor of Medicine at the Case Western Reserve University School of Medicine and also holds appointments in the Departments of Pharmacology and Molecular Biology and Microbiology. Dr. Bonomo's research focuses on microbial resistance to antibiotics, especially β-lactams.
REFERENCES
- 1.Stewardson A, Fankhauser C, De Angelis G, Rohner P, Safran E, Schrenzel J, Pittet D, Harbarth S. 2013. Burden of bloodstream infection caused by extended-spectrum β-lactamase-producing Enterobacteriaceae determined using multistate modeling at a Swiss university hospital and a nationwide predictive model. Infect Control Hosp Epidemiol 34:133–143. doi: 10.1086/669086. [DOI] [PubMed] [Google Scholar]
- 2.Swaminathan M, Sharma S, Poliansky Blash S, Patel G, Banach DB, Phillips M, LaBombardi V, Anderson KF, Kitchel B, Srinivasan A, Calfee DP. 2013. Prevalence and risk factors for acquisition of carbapenem-resistant Enterobacteriaceae in the setting of endemicity. Infect Control Hosp Epidemiol 34:809–817. doi: 10.1086/671270. [DOI] [PubMed] [Google Scholar]
- 3.Peña C, Pujol M, Ardanuy C, Ricart A, Pallares R, Liñares J, Ariza J, Gudiol F. 1998. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum β-lactamases. Antimicrob Agents Chemother 42:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rottier WC, Ammerlaan HSM, Bonten MJM. 2012. Effects of confounders and intermediates on the association of bacteraemia caused by extended-spectrum β-lactamase-producing Enterobacteriaceae and patient outcome: a meta-analysis. J Antimicrob Chemother 67:1311–1320. doi: 10.1093/jac/dks065. [DOI] [PubMed] [Google Scholar]
- 5.Schwaber MJ, Navon-Venezia S, Kaye KS, Ben-Ami R, Schwartz D, Carmeli Y. 2006. Clinical and economic impact of bacteremia with extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 50:1257–1262. doi: 10.1128/AAC.50.4.1257-1262.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SY, Kotapati S, Kuti JL, Nightingale CH, Nicolau DP. 2006. Impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species on clinical outcomes and hospital costs: a matched cohort study. Infect Control Hosp Epidemiol 27:1226–1232. doi: 10.1086/507962. [DOI] [PubMed] [Google Scholar]
- 7.Nordmann P, Mariotte S, Naas T, Labia R, Nicolas MH. 1993. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob Agents Chemother 37:939–946. doi: 10.1128/AAC.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordmann P, Poirel L, Toleman MA, Walsh TR. 2011. Does broad-spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J Antimicrob Chemother 66:689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa TT. 2002. Antimicrobial resistance and aging: beginning of the end of the antibiotic era? J Am Geriatr Soc 50:226–229. doi: 10.1046/j.1532-5415.50.7s.2.x. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization. 2014. Antimicrobial resistance global report on surveillance. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1. Accessed 25 March 2015. [Google Scholar]
- 11.Fournier S, Lepainteur M, Kassis-Chikhani N, Huang M, Brun-Buisson C, Jarlier V, AP-HP Outbreaks Control Group. 2012. Link between carbapenemase-producing Enterobacteria carriage and cross-border exchanges: eight-year surveillance in a large French multihospitals institution. J Travel Med 19:320–323. doi: 10.1111/j.1708-8305.2012.00641.x. [DOI] [PubMed] [Google Scholar]
- 12.Kuenzli E, Jaeger VK, Frei R, Neumayr A, DeCrom S, Haller S, Blum J, Widmer AF, Furrer H, Battegay M, Endimiani A, Hatz C. 2014. High colonization rates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli in Swiss travellers to South Asia—a prospective observational multicentre cohort study looking at epidemiology, microbiology and risk factors. BMC Infect Dis 14:528. doi: 10.1186/1471-2334-14-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markovska R, Schneider I, Ivanova D, Mitov I, Bauernfeind A. 2014. Predominance of IncL/M and IncF plasmid types among CTX-M-ESBL-producing Escherichia coli and Klebsiella pneumoniae in Bulgarian hospitals. APMIS 122:608–615. doi: 10.1111/apm.12204. [DOI] [PubMed] [Google Scholar]
- 14.Thaden JT, Lewis SS, Hazen KC, Huslage K, Fowler VG Jr, Moehring RW, Chen LF, Jones CD, Moore ZS, Sexton DJ, Anderson DJ. 2014. Rising rates of carbapenem-resistant Enterobacteriaceae in community hospitals: a mixed-methods review of epidemiology and microbiology practices in a network of community hospitals in the southeastern United States. Infect Control Hosp Epidemiol 35:978–983. doi: 10.1086/677157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bush K. 2013. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci 1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 16.Bush K. 2013. The ABCD's of β-lactamase nomenclature. J Infect Chemother 19:549–559. doi: 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Pitout JD, Nordmann P. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol 2:501–512. doi: 10.2217/17460913.2.5.501. [DOI] [PubMed] [Google Scholar]
- 18.Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V, European Network on Carbapenemases. 2012. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 18:432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 19.Peirano G, Lascols C, Hackel M, Hoban DJ, Pitout JDD. 2014. Molecular epidemiology of Enterobacteriaceae that produce VIMs and IMPs from the SMART surveillance program. Diagn Microbiol Infect Dis 78:277–281. doi: 10.1016/j.diagmicrobio.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JDD. 2013. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother 57:130–136. doi: 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez F, Duin DV. 2013. Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable patients. Cleve Clin J Med 80:225–233. doi: 10.3949/ccjm.80a.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers in Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby G. β-Lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes. Lahey Clinic, Burlington, MA: http://www.lahey.org/Studies/other.asp#table1. Accessed 3 April 2015. [Google Scholar]
- 24.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Y, Yang J, Ye L, Guo L, Zhao Q, Chen R, Chen Y, Han X, Zhao J, Tian S, Han L. 2014. Characterization of KPC-2-producing Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, and Klebsiella oxytoca isolates from a Chinese hospital. Microb Drug Resist 20:264–269. doi: 10.1089/mdr.2013.0150. [DOI] [PubMed] [Google Scholar]
- 26.Tijet N, Sheth PM, Lastovetska O, Chung C, Patel SN, Melano RG. 2014. Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008-2011. PLoS One 9:e116421. doi: 10.1371/journal.pone.0116421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira PS, Borghi M, Albano RM, Lopes JCO, Silveira MC, Marques EA, Oliveira JCR, Asensi MD, Carvalho-Assef APD. 2015. Coproduction of NDM-1 and KPC-2 in Enterobacter hormaechei from Brazil. Microb Drug Resist 21:234–236. doi: 10.1089/mdr.2014.0171. [DOI] [PubMed] [Google Scholar]
- 28.Kazi M, Drego L, Nikam C, Ajbani K, Soman R, Shetty A, Rodrigues C. 2015. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J Clin Microbiol Infect Dis 34:467–472. doi: 10.1007/s10096-014-2249-x. [DOI] [PubMed] [Google Scholar]
- 29.Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, Hinrichs SH, Rupp ME, Fey PD. 2013. KPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 57:37–41. doi: 10.1128/AAC.01062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castanheira M, Deshpande LM, DiPersio JR, Kang J, Weinstein MP, Jones RN. 2009. First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY Antimicrobial Surveillance Program. J Clin Microbiol 47:4129–4130. doi: 10.1128/JCM.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribeiro VB, Zavascki AP, Nodari CS, Sandri AM, Silva MP, Campos JC, Sampaio JLM, Barth AL. 2012. Detection of blaKPC-2 in a carbapenem-resistant Kluyvera georgiana. J Antimicrob Chemother 67:2776–2777. doi: 10.1093/jac/dks294. [DOI] [PubMed] [Google Scholar]
- 32.Jure MA, Duprilot M, Musa HE, López C, de Castillo MC, Weill FX, Arlet G, Decré D. 2014. Emergence of KPC-2-producing Salmonella enterica serotype Schwarzengrund in Argentina. Antimicrob Agents Chemother 58:6335–6336. doi: 10.1128/AAC.03322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picão RC, Cardoso JP, Campana EH, Nicoletti AG, Petrolini FVB, Assis DM, Juliano L, Gales AC. 2013. The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn Microbiol Infect Dis 76:80–85. doi: 10.1016/j.diagmicrobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Robledo IE, Aquino EE, Vázquez GJ. 2011. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother 55:2968–2970. doi: 10.1128/AAC.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother 52:1413–1418. doi: 10.1128/AAC.01103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez MS, Xie G, Johnson S, Davenport K, van Duin D, Perez F, Bonomo RA, Chain P, Tolmasky ME. 2014. Genome sequences of two carbapenemase-resistant Klebsiella pneumoniae ST258 isolates. Genome Announc 2(3):e00558-14. doi: 10.1128/genomeA.00558-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diago-Navarro E, Chen L, Passet V, Burack S, Ulacia-Hernando A, Kodiyanplakkal RP, Levi MH, Brisse S, Kreiswirth BN, Fries BC. 2014. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J Infect Dis 210:803–813. doi: 10.1093/infdis/jiu157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calfee D, Jenkins SG. 2008. Use of active surveillance cultures to detect asymptomatic colonization with carbapenem-resistant Klebsiella pneumoniae in intensive care unit patients. Infect Control Hosp Epidemiol 29:966–968. doi: 10.1086/590661. [DOI] [PubMed] [Google Scholar]
- 39.Wegner C, Hubner N-O, Gleich S, Thalmaier U, Kruger CM, Kramer A. 2013. One-day point prevalence of emerging bacterial pathogens in a nationwide sample of 62 German hospitals in 2012 and comparison with the results of the one-day point prevalence of 2010. GMS Hyg Infect Control 8:Doc12. doi: 10.3205/dgkh000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagliotti C, Ciccarese V, Sarti M, Giordani S, Barozzi A, Braglia C, Gallerani C, Gargiulo R, Lenzotti G, Manzi O, Martella D, Moro ML. 2013. Active surveillance for asymptomatic carriers of carbapenemase-producing Klebsiella pneumoniae in a hospital setting. J Hosp Infect 83:330–332. doi: 10.1016/j.jhin.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 44.Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 β-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18(31):pii=20549. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549. [DOI] [PubMed]
- 46.Huang T-D, Poirel L, Bogaerts P, Berhin C, Nordmann P, Glupczynski Y. 2014. Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers. J Antimicrob Chemother 69:445–450. doi: 10.1093/jac/dkt367. [DOI] [PubMed] [Google Scholar]
- 47.Carrër A, Poirel L, Yilmaz M, Akan Ö, Feriha AC, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brink AJ, Coetzee J, Corcoran C, Clay CG, Hari-Makkan D, Jacobson RK, Richards GA, Feldman C, Nutt L, van Greune J, Deetlefs JD, Swart K, Devenish L, Poirel L, Nordmann P. 2013. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J Clin Microbiol 51:369–372. doi: 10.1128/JCM.02234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, Sifri CD. 2013. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J Clin Microbiol 51:680–683. doi: 10.1128/JCM.02580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellis C, Chung C, Tijet N, Patel SN, Desjardins M, Melano RG, Toye B. 2013. OXA-48-like carbapenemase-producing Enterobacteriaceae in Ottawa, Canada. Diagn Microbiol Infect Dis 76:399–400. doi: 10.1016/j.diagmicrobio.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 51.Seiffert SN, Perreten V, Johannes S, Droz S, Bodmer T, Endimiani A. 2014. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob Agents Chemother 58:2446–2449. doi: 10.1128/AAC.02417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas CP, Moore LSP, Elamin N, Doumith M, Zhang J, Maharjan S, Warner M, Perry C, Turton JF, Johnstone C, Jepson A, Duncan NDC, Holmes AH, Livermore DM, Woodford N. 2013. Early (2008-2010) hospital outbreak of Klebsiella pneumoniae producing OXA-48 carbapenemase in the UK. Int J Antimicrob Agents 42:531–536. doi: 10.1016/j.ijantimicag.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 53.Dautzenberg MJ, Ossewaarde JM, de Kraker ME, van der Zee A, van Burgh S, de Greeff SC, Bijlmer HA, Grundmann H, Cohen Stuart JW, Fluit AC, Troelstra A, Bonten MJ. 2014. Successful control of a hospital-wide outbreak of OXA-48 producing Enterobacteriaceae in the Netherlands, 2009 to 2011. Euro Surveill 19(9):pii=20723. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20723. [DOI] [PubMed]
- 54.Arana DM, Saez D, García-Hierro P, Bautista V, Fernández-Romero S, Ángel de la Cal M, Alós JI, Oteo J. 2015. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin Microbiol Infect Dis 21:148.e1–148.e4. doi: 10.1016/j.cmi.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 55.Hammoudi D, Ayoub Moubareck C, Aires J, Adaime A, Barakat A, Fayad N, Hakime N, Houmani M, Itani T, Najjar Z, Suleiman M, Sarraf R, Karam Sarkis D. 2014. Countrywide spread of OXA-48 carbapenemase in Lebanon: surveillance and genetic characterization of carbapenem-non-susceptible Enterobacteriaceae in 10 hospitals over a one-year period. Int J Infect Dis 29:139–144. doi: 10.1016/j.ijid.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 56.Österblad M, Kirveskari J, Hakanen AJ, Tissari P, Vaara M, Jalava J. 2012. Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008-11). J Antimicrob Chemother 67:2860–2864. doi: 10.1093/jac/dks299. [DOI] [PubMed] [Google Scholar]
- 57.Mnif B, Ktari S, Chaari A, Medhioub F, Rhimi F, Bouaziz M, Hammami A. 2013. Nosocomial dissemination of Providencia stuartii isolates carrying bla OXA-48, bla PER-1, bla CMY-4 and qnrA6 in a Tunisian hospital. J Antimicrob Chemother 68:329–332. doi: 10.1093/jac/dks386. [DOI] [PubMed] [Google Scholar]
- 58.Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. 2015. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alenton L, Hsann M, Tambyah P, Lin R, Ng K, Rethanam S, Poh L, Soong S, Pada S. 2013. O048: OXA-181-carbapenemase producing Klebsiella pneumoniae. An emerging threat? The first reported nosocomial outbreak in Singapore. Antimicrob Resist Infect Control 2:O48. [Google Scholar]
- 61.Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin Microbiol Infect 18:E144–E148. doi: 10.1111/j.1469-0691.2012.03796.x. [DOI] [PubMed] [Google Scholar]
- 62.Oueslati S, Nordmann P, Poirel L. 2015. Heterogeneous hydrolytic features for OXA-48-like β-lactamases. J Antimicrob Chemother 70:1059–1063. doi: 10.1093/jac/dku524. [DOI] [PubMed] [Google Scholar]
- 63.Perez F, Endimiani A, Ray AJ, Decker BK, Wallace CJ, Hujer KM, Ecker DJ, Adams MD, Toltzis P, Dul MJ, Windau A, Bajaksouzian S, Jacobs MR, Salata RA, Bonomo RA. 2010. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J Antimicrob Chemother 65:1807–1818. doi: 10.1093/jac/dkq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moellering RC. 2010. NDM-1—a cause for worldwide concern. N Engl J Med 363:2377–2379. doi: 10.1056/NEJMp1011715. [DOI] [PubMed] [Google Scholar]
- 65.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 67.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-β-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jain A, Hopkins KL, Turton J, Doumith M, Hill R, Loy R, Meunier D, Pike R, Livermore DM, Woodford N. 2014. NDM carbapenemases in the United Kingdom: an analysis of the first 250 cases. J Antimicrob Chemother 69:1777–1784. doi: 10.1093/jac/dku084. [DOI] [PubMed] [Google Scholar]
- 69.Castanheira M, Deshpande LM, Farrell SE, Shetye S, Shah N, Jones RN. 2013. Update on the prevalence and genetic characterization of NDM-1-producing Enterobacteriaceae in Indian hospitals during 2010. Diagn Microbiol Infect Dis 75:210–213. doi: 10.1016/j.diagmicrobio.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 70.Roy S, Singh AK, Viswanathan R, Nandy RK, Basu S. 2011. Transmission of imipenem resistance determinants during the course of an outbreak of NDM-1 Escherichia coli in a sick newborn care unit. J Antimicrob Chemother 66:2773–2780. doi: 10.1093/jac/dkr376. [DOI] [PubMed] [Google Scholar]
- 71.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chouchani C, Marrakchi R, Henriques I, Correia A. 2013. Occurrence of IMP-8, IMP-10, and IMP-13 metallo-β-lactamases located on class 1 integrons and other extended-spectrum β-lactamases in bacterial isolates from Tunisian rivers. Scand J Infect Dis 45:95–103. doi: 10.3109/00365548.2012.717712. [DOI] [PubMed] [Google Scholar]
- 73.Vergara-López S, Domínguez MC, Conejo MC, Pascual Á, Rodríguez-Baño J. 2013. Wastewater drainage system as an occult reservoir in a protracted clonal outbreak due to metallo-β-lactamase-producing Klebsiella oxytoca. Clin Microbiol Infect 19:E490–E498. doi: 10.1111/1469-0691.12288. [DOI] [PubMed] [Google Scholar]
- 74.Al Bayssari C, Diene SM, Loucif L, Gupta SK, Dabboussi F, Mallat H, Hamze M, Rolain J-M. 2014. Emergence of VIM-2 and IMP-15 carbapenemases and inactivation of oprD gene in carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Lebanon. Antimicrob Agents Chemother 58:4966–4970. doi: 10.1128/AAC.02523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez F, Hujer AM, Marshall SH, Ray AJ, Rather PN, Suwantarat N, Dumford D, O'Shea P, Domitrovic TNJ, Salata RA, Chavda KD, Chen L, Kreiswirth BN, Vila AJ, Haussler S, Jacobs MR, Bonomo RA. 2014. Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in northeast Ohio. Antimicrob Agents Chemother 58:5929–5935. doi: 10.1128/AAC.02372-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lavigne J-P, Sotto A, Nicolas-Chanoine M-H, Bouziges N, Pagès J-M, Davin-Regli A. 2013. An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int J Antimicrob Agents 41:130–136. doi: 10.1016/j.ijantimicag.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 77.Arabestani MR, Rajabpour M, Yousefi Mashouf R, Alikhani MY, Mousavi SM. 2015. Expression of efflux pump MexAB-OprM and OprD of Pseudomonas aeruginosa strains isolated from clinical samples using qRT-PCR. Arch Iran Med 18:102–108. [PubMed] [Google Scholar]
- 78.Moyá B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y, Israeli KPC Kpn Study Group. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother 53:818–820. doi: 10.1128/AAC.00987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adler A, Navon-Venezia S, Moran-Gilad J, Marcos E, Schwartz D, Carmeli Y. 2011. Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J Clin Microbiol 49:2239–2242. doi: 10.1128/JCM.02566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kochar S, Sheard T, Sharma R, Hui A, Tolentino E, Allen G, Landman D, Bratu S, Augenbraun M, Quale J. 2009. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect Control Hosp Epidemiol 30:447–452. doi: 10.1086/596734. [DOI] [PubMed] [Google Scholar]
- 82.Enfield KB, Huq NN, Gosseling MF, Low DJ, Hazen KC, Toney DM, Slitt G, Zapata HJ, Cox HL, Lewis JD, Kundzins JR, Mathers AJ, Sifri CD. 2014. Control of simultaneous outbreaks of carbapenemase-producing Enterobacteriaceae and extensively drug-resistant Acinetobacter baumannii infection in an intensive care unit using interventions promoted in the Centers for Disease Control and Prevention 2012 carbapenemase-resistant Enterobacteriaceae toolkit. Infect Control Hosp Epidemiol 35:810–817. doi: 10.1086/676857. [DOI] [PubMed] [Google Scholar]
- 83.Muñoz-Price L, De La Cuesta C, Adams S, Wyckoff M, Cleary T, McCurdy S, Huband M, Lemmon M, Lescoe M, Dibhajj F, Hayden M, Lolans K, Quinn J. 2010. Successful eradication of a monoclonal strain of Klebsiella pneumoniae during a K. pneumoniae carbapenemase-producing K. pneumoniae outbreak in a surgical intensive care unit in Miami, Florida. Infect Control Hosp Epidemiol 31:1074–1077. doi: 10.1086/656243. [DOI] [PubMed] [Google Scholar]
- 84.Munoz-Price LS, Hayden MK, Lolans K, Won S, Calvert K, Lin M, Stemer A, Weinstein RA. 2010. Successful control of an outbreak of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae at a long-term acute care hospital. Infect Control Hosp Epidemiol 31:341–347. doi: 10.1086/651097. [DOI] [PubMed] [Google Scholar]
- 85.Schwaber MJ, Lev B, Israeli A, Solter E, Smollan G, Rubinovitch B, Shalit I, Carmeli Y. 2011. Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin Infect Dis 52:848–855. doi: 10.1093/cid/cir025. [DOI] [PubMed] [Google Scholar]
- 86.Sypsa V, Psichogiou M, Bouzala G-A, Hadjihannas L, Hatzakis A, Daikos GL. 2012. Transmission dynamics of carbapenemase-producing Klebsiella pneumoniae and anticipated impact of infection control strategies in a surgical unit. PLoS One 7:e41068. doi: 10.1371/journal.pone.0041068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vading M, Samuelsen Ø, Haldorsen B, Sundsfjord AS, Giske CG. 2011. Comparison of disk diffusion, Etest and VITEK2 for detection of carbapenemase-producing Klebsiella pneumoniae with the EUCAST and CLSI breakpoint systems. Clin Microbiol Infect 17:668–674. doi: 10.1111/j.1469-0691.2010.03299.x. [DOI] [PubMed] [Google Scholar]
- 88.Doern CD, Dunne WM Jr, Burnham C-AD. 2011. Detection of Klebsiella pneumoniae carbapenemase (KPC) production in non-Klebsiella pneumoniae Enterobacteriaceae isolates by use of the Phoenix, Vitek 2, and disk diffusion methods. J Clin Microbiol 49:1143–1147. doi: 10.1128/JCM.02163-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, Carey RB, Thompson A, Stocker S, Limbago B, Patel JB. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol 45:2723–2725. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Viau RA, Hujer AM, Marshall SH, Perez F, Hujer KM, Briceño DF, Dul M, Jacobs MR, Grossberg R, Toltzis P, Bonomo RA. 2012. “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in northeast Ohio. Clin Infect Dis 54:1314–1321. doi: 10.1093/cid/cis036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leclercq R, Cantón R, Brown DFJ, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy C-J, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 19:141–160. doi: 10.1111/j.1469-0691.2011.03703.x. [DOI] [PubMed] [Google Scholar]
- 92.CLSI. 2015. Performance standards for antimicrobial susceptibilty testing; twenty-fifth informational supplement. CLSI document M100-S25. CLSI, Wayne, PA. [Google Scholar]
- 93.Livermore DM, Andrews JM, Hawkey PM, Ho P-L, Keness Y, Doi Y, Paterson D, Woodford N. 2012. Are susceptibility tests enough, or should laboratories still seek ESBLs and carbapenemases directly? J Antimicrob Chemother 67:1569– 1577. doi: 10.1093/jac/dks088. [DOI] [PubMed] [Google Scholar]
- 94.Giske CG, Martinez-Martinez L, Canton R, Stefani S, Skov R, Glupczynski Y, Nordmann P, Wooton M, Miriagou V, Skov Simonsen G, Zemlickova H, Cohen-Stuart J, Gniadkowski M. 2013. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. EUCAST, Basel, Switzerland. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf. Accessed 2 April 2015.
- 95.Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother 63:427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cai JC, Zhou HW, Zhang R, Chen G-X. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother 52:2014–2018. doi: 10.1128/AAC.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woodford N, Tierno PM, Young K, Tysall L, Palepou M-FI, Ward E, Painter RE, Suber DF, Shungu D, Silver LL, Inglima K, Kornblum J, Livermore DM. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob Agents Chemother 48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang R, Yang L, Cai JC, Zhou HW, Chen G-X. 2008. High-level carbapenem resistance in a Citrobacter freundii clinical isolate is due to a combination of KPC-2 production and decreased porin expression. J Med Microbiol 57:332–337. doi: 10.1099/jmm.0.47576-0. [DOI] [PubMed] [Google Scholar]
- 100.Hornsey M, Phee L, Woodford N, Turton J, Meunier D, Thomas C, Wareham DW. 2013. Evaluation of three selective chromogenic media, CHROMagar ESBL, CHROMagar CTX-M and CHROMagar KPC, for the detection of Klebsiella pneumoniae producing OXA-48 carbapenemase. J Clin Pathol 66:348–350. doi: 10.1136/jclinpath-2012-201234. [DOI] [PubMed] [Google Scholar]
- 101.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 102.Tenover FC, Kalsi RK, Williams PP, Carey RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE, Hanna B. 2006. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis 12:1209–1213. doi: 10.3201/eid1208.060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Woodford N, Eastaway AT, Ford M, Leanord A, Keane C, Quayle RM, Steer JA, Zhang J, Livermore DM. 2010. Comparison of BD Phoenix, Vitek 2, and MicroScan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J Clin Microbiol 48:2999–3002. doi: 10.1128/JCM.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Day KM, Pike R, Winstanley TG, Lanyon C, Cummings SP, Raza MW, Woodford N, Perry JD. 2013. Use of faropenem as an indicator of carbapenemase activity in the Enterobacteriaceae. J Clin Microbiol 51:1881–1886. doi: 10.1128/JCM.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu F, Ahn C, O'Hara JA, Doi Y. 2014. Faropenem disks for screening of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 52:3501–3502. doi: 10.1128/JCM.02837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Endimiani A, Perez F, Bajaksouzian S, Windau AR, Good CE, Choudhary Y, Hujer AM, Bethel CR, Bonomo RA, Jacobs MR. 2010. Evaluation of updated interpretative criteria for categorizing Klebsiella pneumoniae with reduced carbapenem susceptibility. J Clin Microbiol 48:4417–4425. doi: 10.1128/JCM.02458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. 2009. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol 47:1631–1639. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan Y, Yang H, Pan L, Sun K, Fan H, Lu Y, Shi Y. 2014. Improving the efficiency of the modified Hodge test in KPC-producing Klebsiella pneumoniae isolates by incorporating an EDTA disk. Curr Microbiol 69:47–52. doi: 10.1007/s00284-014-0552-5. [DOI] [PubMed] [Google Scholar]
- 109.Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol 50:477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hirsch EB, Chang K-T, Zucchi PC, Francoeur DN, Ledesma KR, Tam VH, Lasco TM. 2014. An evaluation of multiple phenotypic screening methods for Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae. J Infect Chemother 20:224–227. doi: 10.1016/j.jiac.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 111.Pournaras S, Zarkotou O, Poulou A, Kristo I, Vrioni G, Themeli-Digalaki K, Tsakris A. 2013. A combined disk test for direct differentiation of carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J Clin Microbiol 51:2986–2990. doi: 10.1128/JCM.00901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doi Y, Potoski BA, Adams-Haduch JM, Sidjabat HE, Pasculle AW, Paterson DL. 2008. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type β-lactamase by use of a boronic acid compound. J Clin Microbiol 46:4083–4086. doi: 10.1128/JCM.01408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Porres-Osante N, de Champs C, Dupont H, Torres C, Mammeri H. 2014. Use of avibactam to detect Ambler class A carbapenemases and OXA-48 β-lactamases in Enterobacteriaceae. Diagn Microbiol Infect Dis 79:399–400. doi: 10.1016/j.diagmicrobio.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 114.Huang T-D, Berhin C, Bogaerts P, Glupczynski Y. 2014. Evaluation of avibactam-supplemented combination disk tests for the detection of OXA-48 carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 79:252–254. doi: 10.1016/j.diagmicrobio.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 115.Yagi T, Wachino J, Kurokawa H, Suzuki S, Yamane K, Doi Y, Shibata N, Kato H, Shibayama K, Arakawa Y. 2005. Practical methods using boronic acid compounds for identification of class C β-lactamase-producing Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 43:2551–2558. doi: 10.1128/JCM.43.6.2551-2558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tenover FC, Emery SL, Spiegel CA, Bradford PA, Eells S, Endimiani A, Bonomo RA, McGowan JE. 2009. Identification of plasmid-mediated AmpC beta-lactamases in Escherichia coli, Klebsiella spp., and Proteus species can potentially improve reporting of cephalosporin susceptibility testing results. J Clin Microbiol 47:294–299. doi: 10.1128/JCM.01797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miriagou V, Tzelepi E, Kotsakis SD, Daikos GL, Bou Casals J, Tzouvelekis LS. 2013. Combined disc methods for the detection of KPC- and/or VIM-positive Klebsiella pneumoniae: improving reliability for the double carbapenemase producers. Clin Microbiol Infect 19:E412–E415. doi: 10.1111/1469-0691.12238. [DOI] [PubMed] [Google Scholar]
- 118.Dortet L, Poirel L, Nordmann P. 2012. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clin Microbiol 50:3773–3776. doi: 10.1128/JCM.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dortet L, Brechard L, Poirel L, Nordmann P. 2014. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J Med Microbiol 63:772–776. doi: 10.1099/jmm.0.071340-0. [DOI] [PubMed] [Google Scholar]
- 121.Dortet L, Bréchard L, Poirel L, Nordmann P. 2014. Rapid detection of carbapenemase-producing Enterobacteriaceae from blood cultures. Clin Microbiol Infect 20:340–344. doi: 10.1111/1469-0691.12318. [DOI] [PubMed] [Google Scholar]
- 122.Dortet L, Poirel L, Errera C, Nordmann P. 2014. CarbAcineto NP test for rapid detection of carbapenemase-producers in Acinetobacter spp. J Clin Microbiol 52:2359–2364. doi: 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Österblad M, Hakanen AJ, Jalava J. 2014. Evaluation of the Carba NP test for carbapenemase detection. Antimicrob Agents Chemother 58:7553–7556. doi: 10.1128/AAC.02761-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang T-D, Berhin C, Bogaerts P, Glupczynski Y. 2014. Comparative evaluation of two chromogenic tests for the rapid detection of carbapenemase in Enterobacteriaceae and in Pseudomonas aeruginosa isolates. J Clin Microbiol 52:3060–3063. doi: 10.1128/JCM.00643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pires J, Novais A, Peixe L. 2013. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bernabeu S, Poirel L, Nordmann P. 2012. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn Microbiol Infect Dis 74:88–90. doi: 10.1016/j.diagmicrobio.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 129.Dortet L, Bréchard L, Cuzon G, Poirel L, Nordmann P. 2014. Strategy for rapid detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 58:2441–2445. doi: 10.1128/AAC.01239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel J-M, Drissi M, Mesli E, Touati A, Rolain J-M. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carvalhaes CG, Cayô R, Visconde MF, Barone T, Frigatto EAM, Okamoto D, Assis DM, Juliano L, Machado AMO, Gales AC. 2014. Detection of carbapenemase activity directly from blood culture vials using MALDI-TOF MS: a quick answer for the right decision. J Antimicrob Chemother 69:2132–2136. doi: 10.1093/jac/dku094. [DOI] [PubMed] [Google Scholar]
- 132.Kulkarni MV, Tichy AN, Pyka JS, Murray TS, Hodsdson ME, Peaper DR. 2014. Use of imipenem to detect KPC, NDM, OXA, IMP, and VIM carbapenemase activity from Gram-negative rods in 75 minutes using liquid chromatography-tandem mass spectrometry. J Clin Microbiol 52:2500–2505. doi: 10.1128/JCM.00547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-β-lactamase NDM in Acinetobacter baumannii. J Clin Microbiol 50:1419–1421. doi: 10.1128/JCM.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Swanson DJ, DeAngelis C, Smith IL, Schentag JJ. 1986. Degradation kinetics of imipenem in normal saline and in human serum. Antimicrob Agents Chemother 29:936–937. doi: 10.1128/AAC.29.5.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 136.Mathers AJ, Poulter M, Dirks D, Carroll J, Sifri CD, Hazen KC. 2014. Clinical microbiology costs for methods of active surveillance for Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Infect Control Hosp Epidemiol 35:350–355. doi: 10.1086/675603. [DOI] [PubMed] [Google Scholar]
- 137.Rajapakse N, Vayalumkal J, Lam-Li D, Pearce C, Rees G, Kamhuka L, Peirano G, Pidhorney C, Ledgerwood D, Alfieri N, Hope K, Gregson D, Pitout J, Louie T, Conly J. 2014. Pilot testing of an out-of-country medical care questionnaire with screening and cost analysis of preemptive isolation for carbapenem-resistant Enterobacteriaceae in a large Canadian health region. Infect Control Hosp Epidemiol 35:450–451. doi: 10.1086/675616. [DOI] [PubMed] [Google Scholar]
- 138.CDC. 2008. Laboratory protocol for detection of carbapenem-resistant or carbapenamase-producing, Klebsiella spp. and E. coli from rectal swabs. CDC, Atlanta, GA. http://www.cdc.gov/HAI/pdfs/labSettings/Klebsiella_or_Ecoli.pdf. Accessed 30 March 2015.
- 139.Papadimitriou-Olivgeris M, Bartzavali C, Christofidou M, Bereksi N, Hey J, Zambardi G, Spiliopoulou I. 2014. Performance of chromID CARBA medium for carbapenemases-producing Enterobacteriaceae detection during rectal screening. Eur J Clin Microbiol Infect Dis 33:35–40. doi: 10.1007/s10096-013-1925-6. [DOI] [PubMed] [Google Scholar]
- 140.Vrioni G, Daniil I, Voulgari E, Ranellou K, Koumaki V, Ghirardi S, Kimouli M, Zambardi G, Tsakris A. 2012. Comparative evaluation of a prototype chromogenic medium (ChromID CARBA) for detecting carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J Clin Microbiol 50:1841–1846. doi: 10.1128/JCM.06848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vasoo S, Cunningham SA, Kohner PC, Mandrekar JN, Lolans K, Hayden MK, Patel R. 2013. Rapid and direct real-time detection of blaKPC and blaNDM from surveillance samples. J Clin Microbiol 51:3609–3615. doi: 10.1128/JCM.01731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Griffitt KJ, Grimes DJ. 2013. A novel agar formulation for isolation and direct enumeration of Vibrio vulnificus from oyster tissue. J Microbiol Methods 94:98–102. doi: 10.1016/j.mimet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 143.Girlich D, Poirel L, Nordmann P. 2013. Comparison of the SUPERCARBA, CHROMagar KPC, and Brilliance CRE screening media for detection of Enterobacteriaceae with reduced susceptibility to carbapenems. Diagn Microbiol Infect Dis 75:214–217. doi: 10.1016/j.diagmicrobio.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 144.Wilkinson KM, Winstanley TG, Lanyon C, Cummings SP, Raza MW, Perry JD. 2012. Comparison of four chromogenic culture media for carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 50:3102–3104. doi: 10.1128/JCM.01613-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moran Gilad J, Carmeli Y, Schwartz D, Navon-Venezia S. 2011. Laboratory evaluation of the CHROMagar KPC medium for identification of carbapenem-nonsusceptible Enterobacteriaceae. Diagn Microbiol Infect Dis 70:565–567. doi: 10.1016/j.diagmicrobio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 146.Cohen Stuart J, Voets G, Rottier W, Voskuil S, Scharringa J, Dijk KV, Fluit AC, Hall ML-V. 2013. Evaluation of the Oxoid Brilliance CRE agar for the detection of carbapenemase-producing Enterobacteriaceae. Eur J Clin Microbiol Infect Dis 32:1445–1449. doi: 10.1007/s10096-013-1896-7. [DOI] [PubMed] [Google Scholar]
- 147.Huang T, Berhin C, Glupczynski Y. 2012. Comparative evaluation of the performance of selective media for the detection of carbapenemase-producing Enterobacteriaceae isolates, poster 280. Abstr 32nd Reun Inderdiscip Chimiother Anti Infect, Paris, France. [Google Scholar]
- 148.Giakkoupi P, Tzouvelekis LS, Daikos GL, Miriagou V, Petrikkos G, Legakis NJ, Vatopoulos AC. 2005. Discrepancies and interpretation problems in susceptibility testing of VIM-1-producing Klebsiella pneumoniae isolates. J Clin Microbiol 43:494–496. doi: 10.1128/JCM.43.1.494-496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nordmann P, Girlich D, Poirel L. 2012. Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium. J Clin Microbiol 50:2761–2766. doi: 10.1128/JCM.06477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lolans K, Calvert K, Won S, Clark J, Hayden MK. 2010. Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J Clin Microbiol 48:836–841. doi: 10.1128/JCM.01988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Samra Z, Bahar J, Madar-Shapiro L, Aziz N, Israel S, Bishara J. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 46:3110–3111. doi: 10.1128/JCM.00249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, Mileguir F, Vax M, Ben David D, Tal I, Rahav G, Shamiss A, Mendelson E, Keller N. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J Clin Microbiol 46:2879–2883. doi: 10.1128/JCM.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schechner V, Straus-Robinson K, Schwartz D, Pfeffer I, Tarabeia J, Moskovich R, Chmelnitsky I, Schwaber MJ, Carmeli Y, Navon-Venezia S. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J Clin Microbiol 47:3261–3265. doi: 10.1128/JCM.02368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Landman D, Salvani JK, Bratu S, Quale J. 2005. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J Clin Microbiol 43:5639–5641. doi: 10.1128/JCM.43.11.5639-5641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Singh K, Mangold KA, Wyant K, Schora DM, Voss B, Kaul KL, Hayden MK, Chundi V, Peterson LR. 2012. Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J Clin Microbiol 50:2596–2600. doi: 10.1128/JCM.00654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vasoo S, Lolans K, Li H, Prabaker K, Hayden MK. 2014. Comparison of the CHROMagar KPC, Remel Spectra CRE, and a direct ertapenem disk method for the detection of KPC-producing Enterobacteriaceae from perirectal swabs. Diagn Microbiol Infect Dis 78:356–359. doi: 10.1016/j.diagmicrobio.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 157.Day KM, Ali S, Mirza IA, Sidjabat HE, Silvey A, Lanyon CV, Cummings SP, Abbasi SA, Raza MW, Paterson DL, Perry JD. 2013. Prevalence and molecular characterization of Enterobacteriaceae producing NDM-1 carbapenemase at a military hospital in Pakistan and evaluation of two chromogenic media. Diagn Microbiol Infect Dis 75:187–191. doi: 10.1016/j.diagmicrobio.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 158.Perry JD, Naqvi SH, Mirza IA, Alizai SA, Hussain A, Ghirardi S, Orenga S, Wilkinson K, Woodford N, Zhang J, Livermore DM, Abbasi SA, Raza MW. 2011. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother 66:2288–2294. doi: 10.1093/jac/dkr299. [DOI] [PubMed] [Google Scholar]
- 159.Panagea T, Galani I, Souli M, Adamou P, Antoniadou A, Giamarellou H. 2011. Evaluation of CHROMagar KPC for the detection of carbapenemase-producing Enterobacteriaceae in rectal surveillance cultures. Int J Antimicrob Agents 37:124–128. doi: 10.1016/j.ijantimicag.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 160.Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. 2014. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect 20:350–354. doi: 10.1111/1469-0691.12325. [DOI] [PubMed] [Google Scholar]
- 161.Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PM, Zwinderman AH. 2005. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 162.Doebler P. 2015. mada: meta-analysis of diagnostic accuracy. R package version 0.5.7. R Core Team, Vienna, Austria. [Google Scholar]
- 163.R Core Team. 2014. R: a language and environment for statistical computing. R Core Team, Vienna, Austria. [Google Scholar]
- 164.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 165.Zarakolu P, Day KM, Sidjabat HE, Kamolvit W, Lanyon CV, Cummings SP, Paterson DL, Akova M, Perry JD. 2015. Evaluation of a new chromogenic medium, chromID OXA-48, for recovery of carbapenemase-producing Enterobacteriaceae from patients at a university hospital in Turkey. Eur J Clin Microbiol Infect Dis 34:519–525. doi: 10.1007/s10096-014-2255-z. [DOI] [PubMed] [Google Scholar]
- 166.Bracco S, Migliavacca R, Pini B, Corbo N, Nucleo E, Brigante G, Piazza A, Micheletti P, Luzzaro F. 2013. Evaluation of Brilliance CRE agar for the detection of carbapenem-resistant gram-negative bacteria. New Microbiol 36:181–186. [PubMed] [Google Scholar]
- 167.Girlich D, Anglade C, Zambardi G, Nordmann P. 2013. Comparative evaluation of a novel chromogenic medium (chromID OXA-48) for detection of OXA-48 producing Enterobacteriaceae. Diagn Microbiol Infect Dis 77:296–300. doi: 10.1016/j.diagmicrobio.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 168.Claassen S, du Toit E, Kaba M, Moodley C, Zar HJ, Nicol MP. 2013. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J Microbiol Methods 94:103–110. doi: 10.1016/j.mimet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lupo A, Papp-Wallace KM, Sendi P, Bonomo RA, Endimiani A. 2013. Non-phenotypic tests to detect and characterize antibiotic resistance mechanisms in Enterobacteriaceae. Diagn Microbiol Infect Dis 77:179–194. doi: 10.1016/j.diagmicrobio.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Loman NJ, Constantinidou C, Chan JZM, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. 2012. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 171.Kaase M, Szabados F, Wassill L, Gatermann SG. 2012. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol 50:3115–3118. doi: 10.1128/JCM.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Avlami A, Bekris S, Ganteris G, Kraniotaki E, Malamou-Lada E, Orfanidou M, Paniara O, Pantazatou A, Papagiannitsis CC, Platsouka E, Stefanou I, Tzelepi E, Vagiakou H, Miriagou V. 2010. Detection of metallo-β-lactamase genes in clinical specimens by a commercial multiplex PCR system. J Microbiol Methods 83:185–187. doi: 10.1016/j.mimet.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 173.Voets GM, Fluit AC, Scharringa J, Cohen Stuart J, Leverstein-van Hall MA. 2011. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int J Antimicrob Agents 37:356–359. doi: 10.1016/j.ijantimicag.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 174.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. 2012. Evaluation of a DNA microarray for the rapid detection of extended-spectrum β-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J Antimicrob Chemother 67:1865–1869. doi: 10.1093/jac/dks156. [DOI] [PubMed] [Google Scholar]
- 175.Fishbain JT, Sinyavskiy O, Riederer K, Hujer AM, Bonomo RA. 2012. Detection of extended-spectrum β-lactamase and Klebsiella pneumoniae carbapenemase genes directly from blood cultures by use of a nucleic acid microarray. J Clin Microbiol 50:2901–2904. doi: 10.1128/JCM.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sullivan KV, Deburger B, Roundtree SS, Ventrola CA, Blecker-Shelly DL, Mortensen JE. 2014. Pediatric multicenter evaluation of the Verigene Gram-negative blood culture test for rapid detection of inpatient bacteremia involving Gram-negative organisms, extended-spectrum β-lactamases, and carbapenemases. J Clin Microbiol 52:2416–2421. doi: 10.1128/JCM.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tomita N, Mori Y, Kanda H, Notomi T. 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 178.Liu W, Zou D, Li Y, Wang X, He X, Wei X, Shao C, Li X, Shang W, Yu K, Liu D, Li Y, Guo J, Yin Z, Yuan J. 2012. Sensitive and rapid detection of the New Delhi metallo-β-lactamase gene by loop-mediated isothermal amplification. J Clin Microbiol 50:1580–1585. doi: 10.1128/JCM.06647-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Solanki R, Vanjari L, Ede N, Gungi A, Soory A, Vemu L. 2013. Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of blaNDM-1 and blaKPC genes among carbapenem-resistant clinical Gram-negative isolates. J Med Microbiol 62:1540–1544. doi: 10.1099/jmm.0.059907-0. [DOI] [PubMed] [Google Scholar]
- 180.Ni X, McManus DP, Yan H, Yang J, Lou Z, Li H, Li L, Lei M, Cai J, Fan Y, Li C, Liu Q, Shi W, Liu X, Zheng Y, Fu B, Yang Y, Jia W. 2014. Loop-mediated isothermal amplification (LAMP) assay for the identification of Echinococcus multilocularis infections in canine definitive hosts. Parasit Vectors 7:254. doi: 10.1186/1756-3305-7-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Soli KW, Kas M, Maure T, Umezaki M, Morita A, Siba PM, Greenhill AR, Horwood PF. 2013. Evaluation of colorimetric detection methods for Shigella, Salmonella, and Vibrio cholerae by loop-mediated isothermal amplification. Diagn Microbiol Infect Dis 77:321–323. doi: 10.1016/j.diagmicrobio.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 182.Naas T, Cotellon G, Ergani A, Nordmann P. 2013. Real-time PCR for detection of blaOXA-48 genes from stools. J Antimicrob Chemother 68:101–104. doi: 10.1093/jac/dks340. [DOI] [PubMed] [Google Scholar]
- 183.Tenover FC, Canton R, Kop J, Chan R, Ryan J, Weir F, Ruiz-Garbajosa P, LaBombardi V, Persing DH. 2013. Detection of colonization by carbapenemase-producing Gram-negative bacilli in patients by use of the Xpert MDRO assay. J Clin Microbiol 51:3780–3787. doi: 10.1128/JCM.01092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.van der Zee A, Roorda L, Bosman G, Fluit AC, Hermans M, Smits PH, van der Zanden AG, te Witt R, Bruijnesteijn van Coppenraet LE, Cohen Stuart J, Ossewaarde JM. 2014. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis 14:27. doi: 10.1186/1471-2334-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Babady NE, Gilhuley K, Cianciminio-Bordelon D, Tang Y-W. 2012. Performance characteristics of the Cepheid Xpert vanA assay for rapid identification of patients at high risk for carriage of vancomycin-resistant enterococci. J Clin Microbiol 50:3659–3663. doi: 10.1128/JCM.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Alfa MJ, Sepehri S, Gagne PD, Helawa M, Sandhu G, Harding GKM. 2010. Real-time PCR assay provides reliable assessment of intrapartum carriage of group B Streptococcus. J Clin Microbiol 48:3095–3099. doi: 10.1128/JCM.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Curry SR, Schlackman JL, Hamilton TM, Henderson TK, Brown NT, Marsh JW, Shutt KA, Brooks MM, Pasculle AW, Muto CA, Harrison LH. 2011. Perirectal swab surveillance for Clostridium difficile by use of selective broth preamplification and real-time PCR detection of tcdB. J Clin Microbiol 49:3788–3793. doi: 10.1128/JCM.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lerner A, Romano J, Chmelnitsky I, Navon-Venezia S, Edgar R, Carmeli Y. 2013. Rectal swabs are suitable for quantifying the carriage load of KPC-producing carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 57:1474–1479. doi: 10.1128/AAC.01275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Naas T, Ergani A, Carrer A, Nordmann P. 2011. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob Agents Chemother 55:4038–4043. doi: 10.1128/AAC.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Richter SN, Frasson I, Biasolo MA, Bartolini A, Cavallaro A, Palù G. 2012. Ultrarapid detection of blaKPC1/2-12 from perirectal and nasal swabs by use of real-time PCR. J Clin Microbiol 50:1718–1720. doi: 10.1128/JCM.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nijhuis R, Samuelsen Ø, Savelkoul P, van Zwet A. 2013. Evaluation of a new real-time PCR assay (Check-Direct CPE) for rapid detection of KPC, OXA-48, VIM, and NDM carbapenemases using spiked rectal swabs. Diagn Microbiol Infect Dis 77:316–320. doi: 10.1016/j.diagmicrobio.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 192.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. 2013. Probe ligation and real-time detection of KPC, OXA-48, VIM, IMP, and NDM carbapenemase genes. Diagn Microbiol Infect Dis 76:502–505. doi: 10.1016/j.diagmicrobio.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 193.McEwan AS, Derome A, Meunier D, Burns PJ, Woodford N, Dodgson AR. 2013. Evaluation of the NucliSENS EasyQ KPC assay for detection of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 51:1948–1950. doi: 10.1128/JCM.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.García-Fernández S, Morosini M-I, Marco F, Gijón D, Vergara A, Vila J, Ruiz-Garbajosa P, Cantón R. 2015. Evaluation of the eazyplex SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J Antimicrob Chemother 70:1047–1050. doi: 10.1093/jac/dku476. [DOI] [PubMed] [Google Scholar]
- 195.Vergara A, Zboromyrska Y, Mosqueda N, Morosini MI, García-Fernández S, Roca I, Cantón R, Marco F, Vila J. 2014. Evaluation of a loop-mediated isothermal amplification-based methodology to detect carbapenemase carriage in Acinetobacter clinical isolates. Antimicrob Agents Chemother 58:7538–7540. doi: 10.1128/AAC.03870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 197.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 25th informational supplement update. CLSI M100-S20U. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 198.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 199.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 200.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]