Abstract
Sézary Syndrome is a rare leukemic form of cutaneous T-cell lymphoma defined as erythroderma, adenopathy, and circulating atypical T-lymphocytes. It is rarely curable with poor prognosis. Here we present a multi-platform genomic analysis of 37 Sézary Syndrome patients that implicates dysregulation of the cell cycle checkpoint and T-cell signaling. Frequent somatic alterations were identified in TP53, CARD11, CCR4, PLCG1, CDKN2A, ARID1A, RPS6KA1, and ZEB1. Activating CCR4 and CARD11 mutations were detected in nearly a third of patients. ZEB1, a transcription repressor essential for T-cell differentiation, was deleted in over half of patients. IL32 and IL2RG were over-expressed in nearly all cases. Analysis of T-cell receptor Vβ and Vα expression revealed ongoing rearrangement of the receptors after the expansion of a malignant clone in one third of subjects. Our results demonstrate profound disruption of key signaling pathways in Sézary Syndrome and suggest potential targets for novel therapies.
Cutaneous T-cell lymphomas (CTCL), are heterogeneous lymphoproliferative disorders of clonal skin homing T-cells. Sézary Syndrome (SS), the leukemic variant of Mycosis Fungoides (MF), is defined as erythroderma ( ≥ 80% redness of the skin, adenopathy and ≥1,000/μL circulating Sézary cells with a CD4+CD26– or a CD4+CD7– phenotype1-4. SS cells have a type 2 helper T cell (Th2) cytokine profile1-4. Other features are Staphylococcus aureus colonization, eosinophilia, high IgE levels, chronic, keratoderma with tinea, leonine facies, and secondary malignancies5,6. Depletion of the T-cell repertoire diversity or complexity may cause profound immunosuppression and susceptibility to lethal infection with Cytomegalovirus and S. aureus7,8. SS has a median overall survival of only 2.4 years in patients with >10,000/μL Sézary cells or 5.4 years in patients with 1000–10,000/μL Sézary cells9-12.
MF and SS are the most common CTCLs. SS can arise de novo or it can appear following years of chronic MF1,12, suggesting these two diseases are related at some deeper level. MF and SS are thought to arise from clonal expansion of CD4+ helper T-cells responding to chronic antigen stimulation. Loss of Fas/Fas ligand mediated activation-induced cell death causes accumulation of the clones which may acquire mutations13,14.
Until now, molecular genetic features of SS were limited to small patient cohorts, derived from a single molecular platform. Early cytogenetics and array comparative genomic hybridization (aCGH) approaches15-19 revealed focal deletions of TNFAIP320, CDKN2A21. Mutation of TP5322 reported in a small study, precluded assessment of population frequencies. Recently, targeted sequencing of CTCL patients revealed novel mutations in PLCG1 in about 27% of MF patients and 9% of SS patients23. Genomic gains and point mutations of TNFRSF1B, encoding TNFR2, were recently reported in 18% of MF and SS patients24. Seventeen genes with mutations and loss of tumor suppressors were very recently reported using genomic sequencing with RNA-seq25. PLCG1 and TNFR2 are critical mediators of T-cell receptor (TCR) signaling. Genomic sequencing is shedding new light on the mechanisms of CTCLs and offer new options for targeted therapy.
Although there are several therapies under evaluation that target T-cells, none have led to a cure6,26,27. Hence, our efforts focused on comprehensive, cross-platform integrated analysis of patients with Sézary Syndrome, with attention towards dissecting the molecular pathogenesis and identifying novel clinically actionable molecular targets.
RESULTS
Clinical and histopathological data
A total of 37 SS patients with significant blood involvement (Stage IVA or IVB) were enrolled in the discovery study. All patients provided informed consent for genomic analysis and Institutional Review Boards approved tissue collection. Eleven patients had a preceding history of MF. The clinical and histopathological data were summarized in Supplementary Table 1. CD4+ T-cells were isolated from the peripheral blood of SS patients (Online Methods). We confirmed the identity of Sézary cells as CD4+CD26– cells with a Th-2 phenotype by examining the expression of cell surface markers and Th1/Th2 cytokine expression profiles of the patients (Supplementary Fig. 1). A skin biopsy from each patient's sun-shielded skin was used to culture and expand fibroblasts, from which genomic DNA was extracted and used as matched control for whole-exome sequencing (WES) and SNP array analyses. In addition, CD4+ T-cells were isolated from five healthy donors and their mRNAs were used as controls for whole transcriptome sequencing (RNA-seq). An extension cohort of 68 patients was included and genomic DNAs were extracted from their peripheral blood mononuclear cells (PBMCs). The experimental details were summarized in Supplementary Table 2.
Somatic mutations
WES was performed on tumor and matched fibroblast DNAs as described previously28, yielding a mean coverage of 100×, approximately 94% of targeted bases were covered to a depth of 20× or more. A total of 4,738 somatic mutations were identified (Supplementary Table 3). Validation of mutations in 35 recurrently altered genes was accomplished using a custom capture array (NimbleGen, Inc., Supplementary Table 4) and deep sequencing. The validation rates were 100% for substitutions and 93% for small insertions and deletions (Supplementary Table 5). In addition, for the 32 patients with RNA-seq data available, the mutant alleles of 1,090 somatic mutations were expressed in their mRNAs (Supplementary Table 3 and Supplementary Fig. 2).
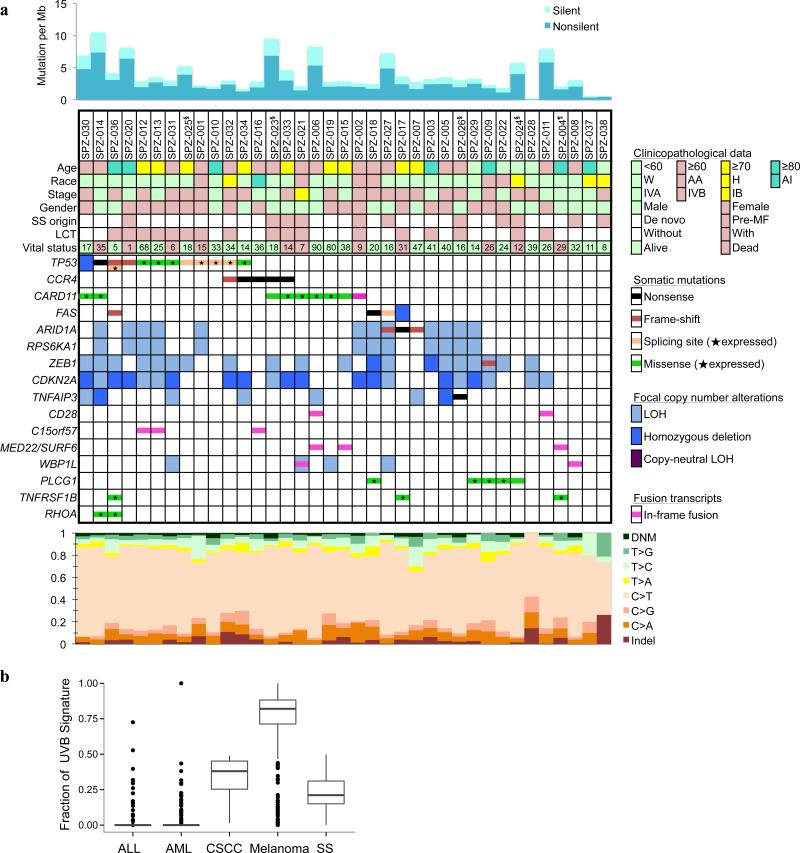
The average somatic mutation rate was 3.85 mutations per megabase (Mb) of targeted DNA and the non-synonymous mutation rate was 2.75 mutations per Mb (Supplementary Fig. 3), which is comparable to adult solid tumors29,30. The most frequent substitution was the cytosine to thymine (C>T) transition (Fig. 1) averaging 74% of mutations across the cohort. As shown in Supplementary Fig. 4a, this high C>T frequency was attributable to two separate mutational processes: 43% of the C>T were at NpCpG sites, which is regarded as age-related31 resulting from spontaneous deamination at CpG sites; 30% of the C>T were at NpCpC sites, T being most prevalent 5’ nucleotide for the signature of UVB exposure31. The fraction of UVB signature in SS is much higher than that of other liquid tumor types (ALL and AML) but is similar to that of cutaneous squamous cell carcinoma (CSCC), see Fig. 1b. Consistent with the prevalence of UVB mutagenesis, a relatively large fraction (2.2%) of dinucleotide mutations (DNM) were found; 68.3% of those were CC>TT. The UVB is a powerful mutagen, which increases the overall mutation frequency. Consistent with this, the signature fractions correlated significantly with overall mutation frequencies (Supplementary Fig. 4b). Prior MF phase has no effect on the amount or percentage of UVB signature observed in a subject (P = 0.15). Although 13 patients received prior focal skin UVB or UVA treatment, no correlation was observed between the presence of UVB signature and the history of therapeutic UV exposure (Supplementary Fig. 4c). This indicates that the accumulated life-time exposure to UV radiation alone explains these UV signatures of SS patients, and is challenging the view that SS originates from the memory T-cells that were in circulation (central memory, Tcm), rather than those resident in the skin (resident memory, Trm)32.
Fig. 1. Somatic genomic alterations identified in SS patients.
a) The upper panel shows somatic mutation rates estimated by WES. The middle panel shows somatic mutations by patient (columns) and by gene (rows) including point mutations, somatic copy number alterations, and recurrent fusion transcripts. The top rows are patient IDs, followed by clinical and histopathological data. Race: W, White; AA, African American; H, Hispanic; AI, American Indian. SS origin: Pre-MF, pre-existing MF. LCT, large cell transformation. LOH, loss of heterozygosity. The number of months lived after disease diagnosis to last clinical visit or death was indicated together with the vital status. Annotation of RNA-seq expression was added for all missense and splicing mutations. A star indicates that the mutant allele was observed in the corresponding transcripts from RNA-seq data. Only genes that met one of the following criteria were included in the table: significantly mutated genes (MutSigCV, false discovery rate <0.1; BCMsig, total points ≥9); significant focal copy number alterations; with in-frame fusions in ≥2 patients; cancer related or biologically important (overlap with Cancer Gene Census or MSigDB database, see URLs) and with 2 or more nonsense, frame-shift, or expressed missense mutations, and were replicated in the extension cohort. Bottom panel: the mutational spectral. DNM, dinucleotide mutations; Indel, small insertions and deletions. §RNA-seq was not performed; ¶SNP array was not performed, due to lack of materials. b) Prevalence of UVB signatures in SS and other cancer types.
Two genes, TP53 and CCR4 were determined to be cancer drivers based on mutation frequency evidence alone in the discovery cohort (Supplementary Table 6). We identified three additional likely driver genes by augmenting mutation frequency with evidence from expression data, functional impact, and known cancer-related genes: ARID1A, CARD11, FAS (Supplementary Table 7). To verify the mutation prevalence of these genes, targeted deep sequencing (1200× fold coverage) of 35 selected genes (Supplementary Table 4) was performed in an extended cohort of 68 SS patients. For genes with hotspot mutations and genes with frequent inactivating mutations, we are able to replicate mutation frequencies in the extension set (Fig. 2a, Supplementary Fig. 5, and Supplementary Table 8). We identified proportionately more PLCG1 mutations in the extension cohort (Supplementary Table 9). The most frequently mutated genes in the combined cohorts were: TP53 (24%), PLCG1 (18%), CARD11 (15%), ARID1A (10%), FAS (10%), CCR4 (7%), RHOA (7%), and TNFRSF1B (6%).
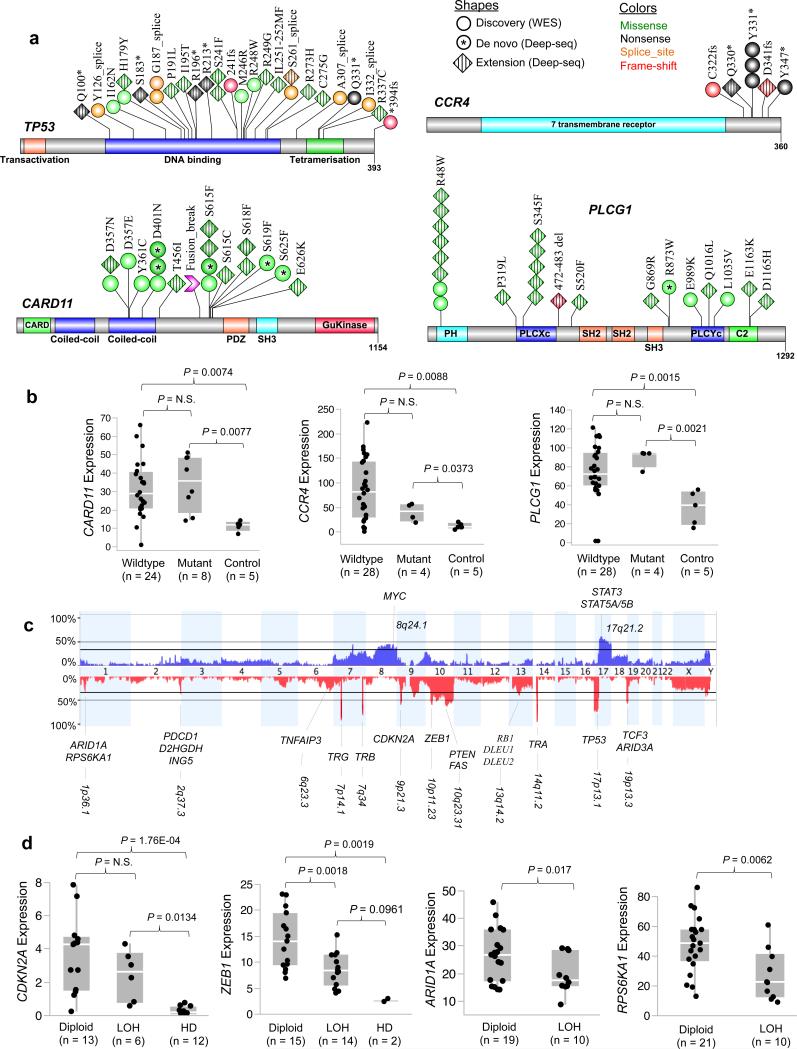
Fig. 2. Significant somatic mutation and copy number alterations.
a) Schematic representation of somatic mutations identified in TP53, CARD11, CCR4, and PLCG1. The red arrowhead indicates the breakpoint for CARD11 fusion; the coiled-coil domains of N-terminal portion are maintained in the fusion. b) Correlation analysis of somatic mutation and gene expression. Those five patients without RNA-seq data were excluded from the analysis. c) Aggregation of somatic copy number alterations. For all frequently occurred focal copy number alterations, the cytogenetic positions were indicated and the names of involved genes that have potential pathogenic significance were displayed; bolded type face were for focal events. For chromosomal arm-level alterations, only the positions and names of well-known cancer-related genes were displayed. d) Correlation analysis of somatic copy number alteration and gene expression. The copy number status was inferred by Nexus (BioDiscovery, Inc.) analysis of the SNP array data. The gene expression levels were normalized FPKM values calculated by the Cufflinks algorithm using RNA-seq data. Those six patients without RNA-seq or SNP array data were excluded from the analysis. For ARID1A correlation analysis, two additional patients with inactivating ARID1A mutations were also excluded. P-values were calculated by one-way ANOVA and student t-test. LOH, loss of heterozygosity; HD, homozygous deletion.
TP53 was the most frequently mutated gene (Fig. 2a). In ten out of eleven patients in the discovery cohort, the somatic mutations were accompanied by copy number losses (del 17p). In the discovery cohort, over half (6/11) of TP53 mutations were located at the canonical splice sites. The mutant alleles of all TP53 splicing mutations were expressed and changes of TP53 exon usage accompanied 4 out of 5 splice-site mutations (data not shown).
The chemokine receptor encoded by CCR4 is important for T-cell migration into skin33,34. Overexpression of CCR4 was reported in SS35 and as a result anti-CCR4 therapy is underway with promising early results in MF/SS patients35,36. Recently, gain-of-function, C-terminal CCR4 mutations were reported in 26% of adult T-cell leukemia/lymphoma (ATLL), a disease caused by human T-cell lymphotropic virus-I37, which like SS also exhibits “skin-homing” pathophysiology. In our SS cohort, 7% of patients harbored CCR4 somatic mutations and all of them were located at the C-terminus including the recurrent Y331X, Q330X, and 3 novel sites that were not previously reported (Fig. 2a). All CCR4 mutations were either nonsense or frameshift. Moreover, the CCR4 mutations were heterozygous and tended to be transcribed at lower levels than patients with wild-type CCR4 (Fig. 2b, P > 0.05 with n = 5 patients), consistent with a functional role as activating mutations.
CARD11, mutated in 15% of SS patients (Figs. 1 and 2a), is a multi-domain adaptor protein required for T cell receptor (TCR)-mediated activation of NF-κB signaling, and is a critical regulator of both T and B cell activation and function38-40. Activating CARD11 mutations were reported in 9.6% of diffuse large B cell lymphoma (DLBCL), virtually all affecting the two coiled-coil domains41,42 located at N-terminal of the protein (Fig. 2a). Introduction of CARD11 mutants of both coiled-coil domains resulted in constitutive NF-κB activation in lines of both T and B origin42. In our combined cohort, we identified 16 CARD11 mutations, half of which were located within the second coiled-coil domain. The remaining was at a new hotspot located off the N-terminal side of the PDZ domain (Fig. 2a). All the mutated positions in this new hotspot are at highly conserved residues (Supplementary Fig. 6). Additionally, a CARD11-PIK3R3 fusion (Supplementary Table 10) was detected in a CARD11-wildtype patient, which includes the new hotspot region but not the N-terminal coiled-coil domains. CARD11 expression was significantly increased in patients with both wildtype and mutant CARD11, including the fusion transcript, in comparison to that of healthy donors (Figure 2b). These data provide strong evidence of a role for CARD11 in SS and therefore therapies targeting CARD11 signaling could benefit SS patients as was previously suggested for CARD11-mutated DLBCL42.
PLCG1, a key mediator in TCR signaling is recently found to be mutated in 27% of MF patients and 9% of SS patients23. Five PLCG1 mutations were identified in the discovery cohort (Fig. 2a), but none were at reported sites. In the extension cohort, the frequency of PLCG1 mutations increased to 21% (14/68), from 14% (5/37) in the original, though not significantly different (P > 0.05, Supplementary Table 9). In addition to the reported hotspot mutation S345F, we identified a new hotspot R48W in the C-terminal of the protein (Fig. 2a and Supplementary Fig. 7). Further correlation analysis with SS origin showed that PLCG1 R48W mutation frequency is significantly higher (P = 0.041) in patients evolved from MF (15%) than those occurred de novo (3%).
Other somatically mutated genes determined to be likely drivers were ARID1A, FAS, TNFRSF1B, and RHOA. As shown in Supplementary Fig. 5 and Supplementary Table 9, recurrent mutation of these genes was replicated in the extension cohort. Most of ARID1A mutations were inactivating, consistent with the usual pattern of ARID1A mutations observed in other cancers. Likewise, most of FAS mutations were inactivating. Given the reported down-regulation of FAS, a pro-apoptotic death receptor43,44, inactivating FAS mutations in 7 patients reinforced their putative role in aiding survival of these malignant cells. TNFRSF1B Thr377Ile mutation was recently reported in 5% of MF/SS patients24. Expression of this mutation in T cells leads to enhance non-canonical NF-κB signaling24. We identified six mutations in this gene; one of them was Thr377Ile and the other five mutations were clustered together, indicating a potential mutation hotspot (Supplementary Fig. 5). Finally, seven missense mutations with likely functional impact were observed in RHOA, a small GTPase that acts to regulate actin cytoskeleton and cell division.
Somatic copy number alterations
Somatic DNA copy number analysis revealed frequent genomic gains and losses (Fig. 2c). We identified CDKN2A focal deletion in 58% of the patients, compared to about 40% previously reported21,25. CDKN2A expression was significantly decreased in patients with homozygous deletions (Fig. 2d), as expected.
The most frequent novel event in our discovery cohort was a 0.6-Mb focal deletion at 10p11.23 including ZEB1 and ZNF438 observed in 56% of the patients (Fig. 2c). Of the two genes, only ZEB1 expression was decreased in proportion to copy number (Figure 2d), compared to patients with diploid ZEB1 (P < 0.001), suggesting that ZEB1 may be the target of selection within this focal deletion. ZEB1, also known as T-cell transcription factor 8, is a zinc finger transcriptional repressor that harbors two independent repressor domains; the N-terminal one blocks the activity of hematopoietic transcription factors such as ets and c-myb and regulates hematopoietic differentiation45. ZEB1 has been suggested to be critical for early T-cell development46 and has a role in enhancing transforming growth factor-β1 (TGF- β1) signaling by binding phosphorylated Smad3 and Smad747. ZEB1 downregulation contributes to resistance to TGF-β1-mediated growth suppression in ATLL-derived cell lines47,48. Loss of ZEB1 may play a similar role in SS pathogenesis.
Hemizygous deletion of 1p36.1 (33%) encompassed 6 genes including ARID1A and RPS6KA1. Expression levels of both ARID1A and RPS6KA1 decreased and correlated significantly with their copy number status (Figure 2d), suggesting selection operating on these two genes. As mentioned above, six patients in our combined cohorts harbored inactivating mutations in ARID1A (Supplementary Fig. 5), consistent with the function of ARID1A as an epigenetic tumor suppressor. Although ARID1A is best known as a component of the SWI/SNF chromatin remodeling complex, recent functional studies suggested a role of ARID1A in cell cycle checkpoint machinery, possibly through p53-and/or pRb-dependent signaling cascades49. ARID1A is required for cell cycle exit, since in the absence of functional ARID1A, cell cycle arrest is delayed50. RPS6KA1 is a serine/threonine kinase that plays a pivotal role in growth factor induced G2/M transition delay and is required to maintain genomic integrity during growth factor stimulation51 Thus we place these two genes in an ensemble of mutated genes, the dysregulated replication cycle in Sézary cells (Fig. 3a).
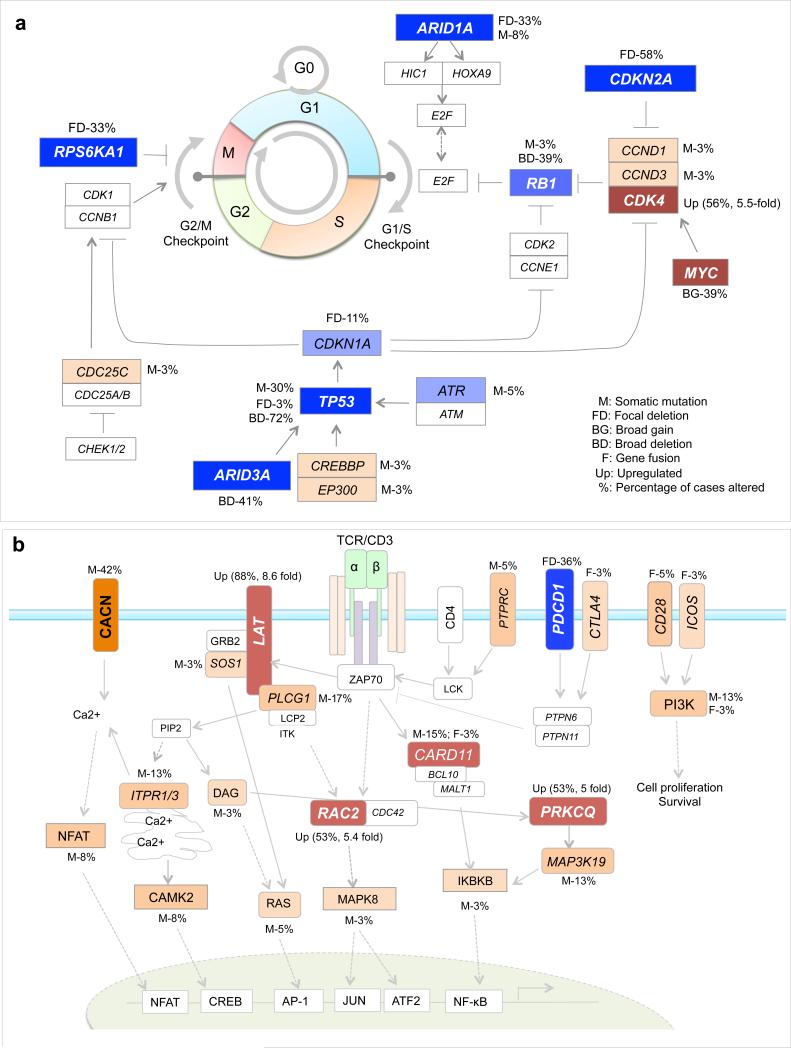
Fig. 3. The dysregulated signaling pathways.
a) The cell cycle checkpoint machinery. b) T-cell receptor (TCR) signaling pathway. The genes with inactivating mutations or copy number losses were colored in blue; the genes with activating mutations, copy number gains or significantly upregulated genes in comparison with control T cells were colored in red; genes with all other genetic alterations were colored in orange; genes without detectable alterations but are important components of the pathway are also included and shown in white rectangles; for each gene with somatic alterations, the percentage of cases altered was indicated; for significantly upregulated genes, the average fold changes are also shown.
Finally, focal deletion of 2q37.3 involving PDCD1 and 5 other genes in 36% of patients (Figure 2c). Subjects with 2q37.3 deletion tend to have a worse prognosis (P = 0.05), see Supplementary Fig. 8. PDCD1 is expressed on activated T cells and transduces a negative signal that suppresses T-cell function52. PDCD1 is suggested to be a promising immunotherapeutic target in cancer since blockage of PDCD1 and its ligands can enhance functioning of cytotoxic T cells53. However, we observed weak correlation between expression and copy number for PDCD1, or any of the 5 known genes in the region (Supplementary Fig. 9).
Fusion transcripts and differentially expressed genes
We identified 41 in-frame fusion transcripts by RNA-seq (Supplementary Table 10). For 29 fusion transcripts with RNAs available, we performed RT-PCR (Supplementary Table 11), sequenced the RT-PCR products and successfully validated all of them. In addition to the aforementioned CARD11-PIK3R3, CD28-CTLA4/ICOS, and IKZF2-GLI2 fusions were observed. CD28 is essential for T-cell proliferation and survival. Low frequency of CD28-CTLA4 fusion was recently reported in CTCL24. IKZF2 regulates lymphocyte development. Overall, gene-gene fusions appeared to be relatively rare in SS pathogenesis.
To identify differentially expressed genes, we performed an unbiased analysis of the expression data for all detectable transcripts (> 40,000). A total of 345 transcripts were significantly upregulated in SS patients (Supplementary Table 12). Gene set enrichment analysis of these genes suggested significant enrichment of only a few canonical pathways (Supplementary Table 13), among which were cell cycle control, regulation of immune system, MYC transcriptional activation, and chemokine signaling.
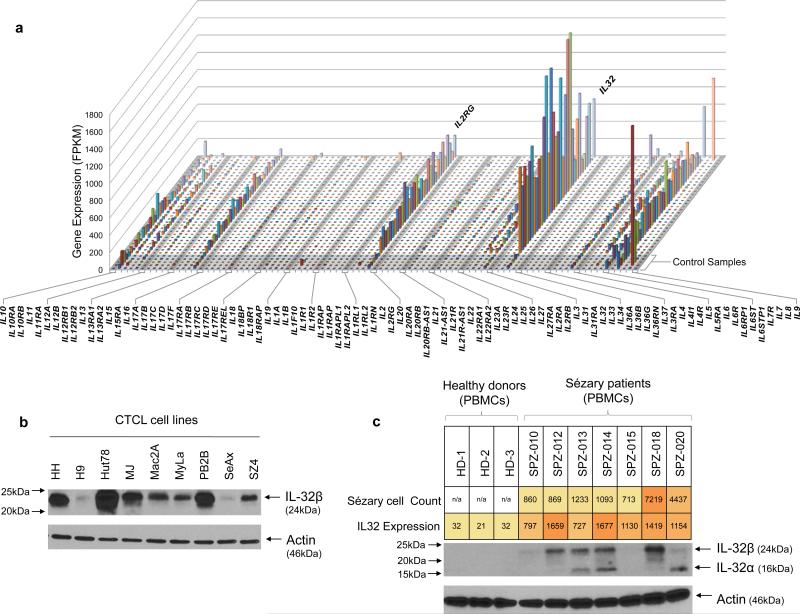
Among those 345 genes, several interleukins (ILs), interleukin receptors, CD molecules, and chemokines that are required for T-cell development and function were strongly upregulated including IL32, IL2RG, CD3G, CD27, CCR4, and CCR8 (Figure 4a). IL2RG, encoding the IL2 receptor common gamma chain, was highly expressed in all patients. IL2RG heterodimerizes with several other receptors and may transmit enhanced cytokine signaling to Sézary cells. IL32 was the most upregulated chemokine (Figure 4b); and strongly upregulated in all but one patient, who had a low level of Sézary cells. Interestingly, IL32 expression levels were positively correlated with those of IL2RG and CARD11 (Supplementary Fig. 10a,b). IL32 is a pro-inflammatory cytokine expressed by activated T cells and natural killer cells with an important role in T-cell communication, tumorigenesis and autoimmune disease54. IL32 accelerates the proliferation of CTCL cell lines through mitogen-activated protein kinase and NF-κB-mediated signaling54. A high level of IL32 expression in skin lesions of MF patients was reported to correlate with disease activity54,55.
Fig. 4. Increased IL32 gene and protein expression.
a) Expression profiles of all the interleukins in SS patients. The gene expression data were generated from RNA-seq. The five control samples were obtained from sorted CD4+ T cells from healthy donors. (b) The cellular IL32 protein expression in 9 CTCL cell lines. (c) IL32 protein expression in the peripheral blood mononuclear cells (PBMCs) from 3 healthy donors (HD-1, 2, 3) and 7 SS patients. The protein expression was assessed by western blot with anti-human IL32 monoclonal antibody. The overall IL32 gene expression levels estimated from RNA-seq data (on purified SS cells) and the number of circulating SS cells in patients’ PBMCs were shown and color scaled.
We examined IL32 protein in CTCL cell lines and tumor cells from a subset of SS patients. IL32α was not detectable in the serum of SS patients or the cell culture supernate of any cell lines (data not shown), suggesting that IL32α might function intracellularly rather than intercellularly in the context of SS cells. Western blot employing a purified anti-human IL32 monoclonal antibody, which has been shown to bind to all isoforms of IL3256, showed a strong single 24-kDa± band, indicative of IL32 β translation in all cell lines (Fig. 4c). IL32 protein expression in patients’ cells was more heterogeneous exhibiting both the α and β isoforms in some patients (Fig. 4d). Overall, the protein expression is correlated with their mRNA expression and the density of Sézary cells in their blood. These data support the possibility that IL32 is involved in an autocrine signaling loop stimulating growth of Sézary cells as suggested by Suga and colleagues54.
Dysregulated signaling pathways
Our integrated genomic analyses revealed frequent alterations primarily in three oncogenic pathways (Supplementary Table 14): epigenetic regulation (95%), TCR signaling (84%) and cell cycle control (78%). Importantly, we identified frequent alterations in genes involved in cell cycle control, especially genes that regulate G1/S and G2/M checkpoints (Fig. 3a). Notably, multiple genes were recurrently altered at a high frequency, including the aforementioned TP53, CDKN2A, ARID1A and RPS6KA1, suggesting that loss of mitotic checkpoint control might be an important contributor to the high rate of genomic instability observed in SS patients.
Additionally, we identified somatic alterations in TCR signaling in 84% of cases (Fig. 3b). TCR signaling in response to antigen recognition has a critical role in the adaptive immune response. Recurrent alterations were identified in CARD11, PLCG1, LAT, RAC2, PRKCQ, CD28, and genes that encode the calcium channel subunits. Finally, though the epigenetic regulation was most frequently altered, the majority of involved genes were mutated only once each. As shown in Supplementary Tables 15-16, all eight classes of epigenetic genes57 were somatically altered across the cohort with the highest frequency (66%) among the histone writers.
Evolution of the SS TCR repertoire
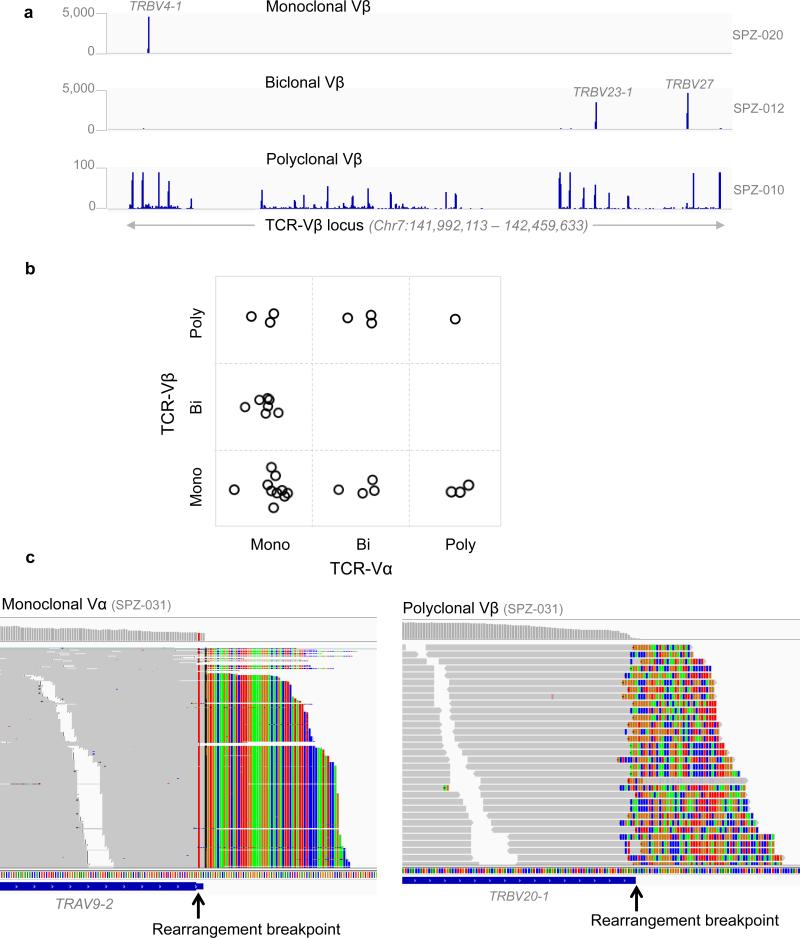
The orderly rearrangement and assembly of TCR genes during T-cell development creates a polyclonal TCR repertoire that is highly diverse in healthy individuals. An important diagnostic correlate in CTCL is the identification of a unique TCR rearrangement pattern present in the circulating malignant T-cells, indicative of an expanded T-cell clone58. With RNA-seq data, we were able to assess all V chains simultaneously. All patients’ TCR expressed Vα and Vβ and two of them also expressed Vγ and Vδ. We detected one or two dominant Vβ- or Vα-expressing clone(s) in 97% of patients (Supplementary Table 17), compared to 67% by flow cytometric analysis of TCR-Vβ using fluorescence-labeled monoclonal antibodies.
Our analysis revealed varied patterns in the dominant Vβ and Vα receptors (Fig. 5a,c). We detected mono-, bi-, and polyclonal TCR-Vβ in 56%, 22%, and 22% of patients respectively, and mono-, bi-, polyclonal TCR-Vα in 66%, 22%, and 12% of patients, respectively (Supplementary Table 17). The most frequent clonally expanded TCR-Vα gene was TRAV27, followed by TRAV9-2. The most frequent clonally expanded TCR-Vβ gene was TRBV20-1, followed by TRBV7-3 (Supplementary Fig. 11). TRBV20-1 is associated with S. aureus infection59, a common feature of SS, and a finding consistent with the antigen-stimulation theory of CTCL60.
Fig. 5. Characterization of TCR clonality using RNA-seq.
(a) Representative RNA-seq read coverage data showing the monoclonal, biclonal and polyclonal TCR-Vβ. The data ranges are shown on the left and the sample IDs are shown on the right. Clonally expressed TCR genes were indicated. b) The relationship of TCR-Vβ and Vα. Bi, biclonal; Mono, monoclonal; poly, polyclonal. Each circle represents a patient, which is placed in a box to indicate the state of its Vα and Vβ expression. c) Representative images showing partially aligned sequencing reads at the breakpoints of a TCR V-region rearrangement for a patient with monoclonal Vα and polyclonal Vβ. The matched sequencing bases are in gray and the non-aligned bases shown color-coded (A, T, C, G; green, red, blue, brown, respectively) indicating the recombination break point where the V region adjoins its partner J region (Vα) or D region (Vβ). At the Vα exon all non-aligned reads exhibit the same sequence (note vertical striping of colors) indicating clonality in expression; at Vβ all non-aligned reads are different (note checkered pattern of colors) indicating polyclonal rearrangements of this V region.
The TCR is thought to be formed by differentiation of a T-cell precursor in the thymus61,62 in which, critically, the Vβ locus is rearranged and expressed on the cell surface, followed by rearrangement of the Vα locus. The T-cells with fully formed, heterodimerized TCR exit the thymus and live in various body compartments until stimulated by antigen. Upon malignant transformation, this simple model of TCR differentiation should lead to a clonal population expressing a single Vα and a single Vβ. In our cohort, 11 out of 32 patients fit the model of a single Vα and Vβ transcript expressed. Interestingly, 6 patients exhibited monoclonal Vβ paired with either a biclonal or poly-clonal Va (Fig. 5b). Clearly, at the point of malignant transformation, these cells were able to further differentiate their Vα chain to produce a second Vα receptor (bi-clonal) or even a fully polyclonal Vα repertoire, each of which is paired to a single (clonal) Vβ. This final differentiation step occurs after the malignant transformation event because the malignant cells are clonal with respect to their mutation profile.
Moreover, an additional 7 patients with monoclonal Vα were biclonal or polyclonal in Vβ. Since the canonical differentiation of the TCR stipulates the Vβ rearrangement occurs before the Va61,62, it is hard to explain how a malignant clone can be monoclonal for Vα and polyclonal for Vβ. One possibility is that some T-cells rearrange the Vα receptor first, contrary to the accepted TCR differentiation pathway; or, alternatively, upon malignant transformation, the initially rearranged Vβ is lost, and a new round of Vβ rearrangement occurs as the malignant cells divide to produce polyclonal Vβ. This latter mechanism would be the simplest one to explain the poly clonality in both the Vα and Vβ. Together, these data suggest greater phenotypic complexity in the display of TCR components than had been previously appreciated.
Correlation and survival analysis
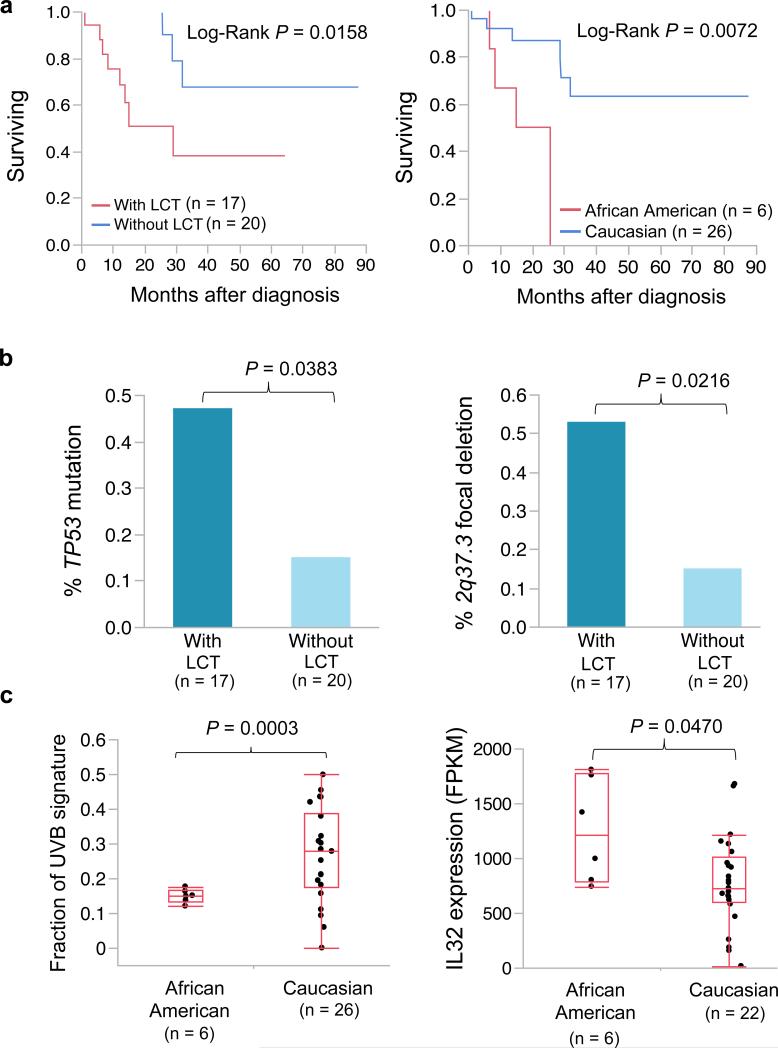
Finally, our correlation and survival analyses confirmed that large cell transformation (LCT) and African ancestry were associated with worse prognosis11,63 (Fig. 6a), which is also replicated in the extension cohort (Supplementary Fig. 12). TP53 mutation and 2q37.3 focal deletion were significantly enriched in patients with LCT (Fig. 6b). Interestingly, we found that prevalence of UVB signature was significantly lower in African American patients while IL32 expression was significantly higher (Figure 6c). Overexpression of IL32 in African American patients with worse prognosis further suggesting a connection between IL32 and the pathophysiology of SS. Our survival analysis also suggested a trend towards worse prognosis in patients with CCR4 alteration, CDKN2A, 2q37.3 deletion (Supplementary Fig. 9).
Fig. 6. Survival and correlation analysis.
For survival analysis (a), the Kaplan-Meier survival plots were shown and the p-values were calculated by Log-Rank test. For frequency comparison (b), the p values were calculated by Fisher's exact test. For comparison of UVB signature and IL32 expression, the p-values were calculated by t-test. Those four Caucasian patients without RNA-seq data were excluded from analysis.
DISCUSSION
To gain more insight into the genomic basis of SS, we assessed the somatic mutation, DNA copy number and gene expression profiles in 37 patients. In addition to somatic alterations in previously known SS-associated genes such TP53 and CDKN2A, novel recurrent mutations and/or focal deletions were identified in CARD11, CCR4, PLCG1, ARID1A, and ZEB1, etc. Several somatic alterations are reported to specifically impact T-cell signaling and differentiation. These data coupled with overexpression of IL32 andIL2RG, lend the first mechanistic insights into the nature of SS pathogenesis, and suggest avenues for development of targeted therapies. Importantly, this study has drawn a number of parallels, through shared molecular features, to other malignancies of both B- and T-cell origin. The CCR4 activating mutations, first reported in ATLL patients, are in common with our SS patients’ mutations and highlight the importance of CCR4 in pathogenesis and targeted therapy of both lymphomas. The mutation of the coiled-coil domain of CARD11 that potentiates NF-κB signaling in B cells has been recognized as a potential therapeutic target in DLBCL. To these markers, SS adds IL32 as possible constituents of an autocrine signaling network, which could become targets specific to SS.
Important to the etiology of the disease is a description of the cell of origin of this malignancy and both Tcm and Trm cells have been proposed32. The surprising finding of a UVB signature in 30% of the patients is strong evidence that the malignant clonal population arose from a cell that spent a significant time in the superficial layers of the skin, consistent with a Trm origin.
Our data provide evidence of disruption of the precise choreography of TCR maturation, which classically proceeds from Vβ to Vα through multiple rounds of cell division. Production of the same Vα rearrangement in many different Vβ-rearranged cells is difficult to explain by the classical Vβ -> Vα pathway. A mechanism is needed to explain how a cell which presumably exited the thymus with intact TCR-Vα and Vβ heterodimer, restarted the rearrangement of either Vα or Vβ to produce a polyclonal TCR. Thus, the genomic profiling presented here not only added to our understanding of genetic driver mutations, T-cell signaling, but also to the pathophysiology of SS and identified novel targets and pathways for developing more effective therapies.
ONLINE METHODS
Patients
Thirty-seven SS patients from the University of Texas MD Anderson Cancer Center who signed consent were approved by the institutional review board to enroll in this study. It was conducted according to the Declaration of Helsinki Principles. The revised diagnostic criteria for staging of SS by ISCL/EORTC were used4.
Isolation of CD4+ cells
Peripheral blood was collected prior to therapy from all SS patients using BD Vacutainer® Cell Preparation Tube. Peripheral blood mononuclear cells (PBMCs) were then isolated from granulocytes. CD4+ T-cells were separated from CD4– lymphocytes using CD4+ T-Cell Isolation Kit (Miltenyi Biotec). The purity and Vβ usage of CD3+CD4+CD26– T-cells were confirmed by flow cytometry; all CD4+ T-cell samples had a purity of > 90%. The CD4– cells were also collected, from which DNAs were isolated and used as a second matched control for identification of somatic mutations. Our analysis suggested contamination of Sézary cells in the CD4– cell population (data not shown) and thus matched fibroblast DNAs were used as germline controls for somatic mutation and copy number analyses.
Skin punch biopsy and fibroblast cell culture
A 6-mm biopsy of normal skin from the inner arm was performed for all patients. Explanted fibroblasts were grown and maintained in Dulbecco's MEM supplemented with non-essential amino acids, 1% penicillin/streptomycin, 1% HEPES, 1% sodium pyruvate, and 10% fetal bovine serum (DMEM-FBS) at 37°C in humidified air containing 5% CO2. Fibroblasts were subcultured using trypsin/EDTA. DNA and total RNA was extracted at passage 3 to 464.
DNA/RNA extraction
For all samples, about 5×106 cells were collected and genomic DNA and total RNA were extracted simultaneously using All Prep DNA/RNA/Protein Mini Kit (QIAGEN) following the manufacturer's protocol.
WES library preparation, exome capture and sequencing
DNA samples were constructed into Illumina paired-end (PE) pre-capture libraries as previously described28 with minor modifications. Briefly, 0.5 μg DNA was sheared into fragments of 200–300 base pairs in a Covaris plate with E210 system (Covaris) followed by end-repair, A-tailing and ligation of Illumina multiplexing PE adaptors. Pre-capture Ligation Mediated-PCR (LM-PCR) was performed using the Library Amplification Readymix containing KAPA HiFi DNA Polymerase (Kapa Biosystems, Cat. No. KK2612). Universal primer IMUX-P1.0 and IMUX-P3.0 were used in PCR amplification. For a sub-set of the libraries constructed in this study, these molecular barcodes were introduced at PCR step using Phusion PCR Supermix HiFi (2X) (NEB, Cat. No. M0531L). Purification was performed with Agencourt AMPure XP beads after enzymatic reactions. Following the final XP beads purification, quantification and size distribution of the pre-capture LMPCR product was determined using the LabChip GX electrophoresis system (PerkinElmer) and gel analysis using AlphaView SA v3.4 software.
For the hybridization step, four such pre-capture libraries were pooled together. The pooled libraries were then hybridized in solution to the HGSC VCRome 2.1 design. For post-capture LM-PCR amplification, either matching KAPA HiFi DNA Polymerase or Phusion PCR Supermix HiFi (2×) was used. After the final AMPure XP bead purification, quantity and size of the capture library was analyzed using the Caliper LabChip GX electrophoresis system. The efficiency of the capture was evaluated by performing a qPCR-based quality check on the enrichment level of four standard NimbleGen control loci. All sequencing runs were performed in PE mode on Illumina HiSeq2000 platform.
WES data processing, mutation calling and filtering
WES data processing, mutation calling and annotation was performed using the standard pipelines established at HGSC as described previously28. A stepwise filtering approach was performed to identify high-confident somatic mutations. Briefly, the quality filters we first applied and only those meeting the following criteria were selected: 1) the total read coverage in tumor and its matched control sample ≥ 5 and 2, respectively; 2) the variant read coverage in tumor ≥ 2; 3) the variant allele fraction (VAF) in tumor ≥ 0.04; 4) the ratio of VAF in normal and its tumor ≤ 0.15. Secondly, the germline filters were applied and any variants observed in the germline MAF file of the cohort were removed. After that, common SNPs were filtered out using dbSNP139 and virtual normals. Finally, all mutations left were visualized in Integrative Genomics Viewer (IGV, see URLs) and the mapping quality and uniqueness of the supporting reads was assessed using BLAT (see URLs). The ones with ambiguous mapping and poor mapping quality were removed from the list.
Mutation significance
The statistical significance of mutation frequency in each gene was determined using the MutSigCV algorithm (v1.4)65 and our in-house BCMsig method. For MutSigCV analysis, a p value of < 0.05 and a q value (false discovery rate) of < 0.1 were considered statistically significant. Given that application of MutSigCV to our dataset revealed only two significantly mutated genes, we employed an alternative approach that is more sensitive to SMG identification for smaller tumor datasets. BCMsig is a simple binary algorithm that borrows elements from MutSigCV, but also incorporates other parameters, such as RNA expression data in both normal T-cells and SS cells, the cancer mutation literature, and likely functional impact of individual mutations. Analysis of the final somatic mutation file for 37 SS patients revealed 480 genes that had at least two non-silent somatic mutations. For each of these genes 13 parameters were analyzed and given scores of −1, 0, or +1, as indicated in rows 1-4 in Supplementary Table 7. Scores for each of the 13 parameters were added up and given a total score. All genes with a total score equal to or greater than that of TP53 (9 points) were considered to be significantly mutated.
Mutation signatures
Non-negative matrix factorization (NMF) was used to evaluate the mutation signatures associated with SS malignant cells, of which there was a set of 21 available. Mutation data were transformed into a data matrix V96,n by normalizing and grouping mutations for each of the 16 mutation contexts (and 6 possibly base changes) for each of n subjects. Solutions were generated by solving the equation: V = W × H + E where W96,21 is the basis matrix of the NMF (Covington and Wheeler in preparation) solution and H21,n is the coefficient matrix for the set of SS subjects, subject to minimizing the squared error (E). H was taken as the set of overall signature exposure values, commonly referred to as the coefficient matrix. H can be taken to mean the number of mutations in a patient's sample attributable to each signature (see Supplementary Fig. 4a and b). H was also scaled such that each column of H summed to 1 to represent the proportion of mutations attributable to each signature (see Figure 1b). Both scaled and unscaled exposure values were merged with clinical, genomic, and transcritpomic data for further analysis.
Mutation validation and mutation prevalence
Thirty-five frequently altered genes were selected and a custom NimbleGen array (target size: ~200kb, Supplementary Table 4) was designed for mutation validation and mutation frequency screening. A total of 142 DNA samples were collected from patients in our combined cohorts. 0.1–0.5 μg DNA per sample was used for validation study.
The basic Illumina library preparation protocol and the regents used were identical to that of WES except that for validation, a set of 96 molecular barcodes were used to allow high density multiplexing. The capture enrichment protocol was also same as described earlier for WES, with the following modifications: for capture, four pools of 35–36 libraries/pool were prepared and hybridized in solution to the custom capture design. Overnight hybridization at 56°C was carried out instead of the 3-day hybridization at 47°C as done for WES. Post enrichment steps were also similar: four enriched pools were pooled into two groups (70 and 72 libraries/pool) and sequenced on two lanes of HiSeq 2500. On an average, 500 Mb of uniquely mapped sequence data were generated across the target region and the targeted bases were covered to 1226×.
For paired samples, the workflow of data processing was similar to that of WES. The mutation is valid if 5 or more supporting reads were detected. For tumor only samples, a stepwise filtering approach was applied to identify high-confident potential somatic mutations. Briefly, only non-silent mutations with ≥ 5 supporting reads and a VAF of ≥ 0.01 were selected, the common SNPs were eliminated using dbSNP database and virtual normals. The list was further cleaned by removing variants present in any of the fibroblast controls. Finally, all left mutations were visualized in IGV and the mapping quality and uniqueness of the supporting reads was assessed using BLAT. The ones with ambiguous mapping and poor mapping quality were removed from the list.
RNA-seq library construction, sequencing, and quality control
High quality total RNA (RIN > 7) was used to prepare strand-specific, poly-A+ Illumina RNA-seq libraries. To monitor sample and process variability, in addition to sequence metrics, RNA developed by the External RNA Controls Consortium (ERCC) which consists of a mix of 92 polyadenylated RNA transcripts was also added to total RNA at the beginning of the library preparation. Briefly, poly-A+ mRNA was extracted from 1 μg total RNA using Oligo(dT)25 Dynabeads (Life Technologies, Cat. No. 61002) followed by fragmentation, first strand cDNA synthesis, second strand cDNA synthesis, end-repair, A-tailing, ligated with Illumina PE adapters, and then digested with 10 units of Uracil-DNA Glycosylase (NEB, Cat. No. M0280L). Amplification of the libraries was performed for 13–14 PCR cycles using the Phusion High-Fidelity PCR Master Mix; twelve such 6-bp molecular barcodes were also incorporated during this PCR amplification. The cDNA was purified with Agencourt AMPure XP beads after each enzymatic reaction, and after quantification using the Agilent Bioanalyzer 2100 DNA Chip 7500 (Cat. No. 5067-1506). Libraries were pooled in equimolar amounts (2 – 5 libraries/pool). The library pools were then loaded onto a HiSeq flowcell lane, and following amplification with Illumina's cBot cluster generation system, sequencing runs were performed in PE mode (2×101 bp reads) using the Illumina HiSeq2000 and HiSeq 2500 platforms. The data generated was processed through an automated RNA-Seq pipeline built for quality control, gene fusion and expression analysis. RNA-SeQC (v1.1.7)66 was applied to generate a series of quality control metrics for RNA-seq data.
Expression quantification and differential gene expression
The FASTQ data generated from production pipeline were mapped to human genome (both hg19 and known splice junction loci) using the STAR RNA-seq aligner (v2.3.1)67. The human genome and gene models (GRCh37.69) were downloaded from Ensembl (see URLs). The transcripts were then assembled with Cufflinks (v2.1.1)68 and the expression level of each gene was calculated and normalized.
Gene fusion detection and filtering
To identify gene fusion transcripts, RNA-seq reads in FASTQ format were aligned to human genome by deFuse algorithm (v0.6.1)69. DeFuse was run with default parameters and a probability score was assigned to each candidate with DeFuse classifier. The candidate gene fusion transcripts were filtered step by step to identify the high-confident fusion events. Briefly, the fusion transcripts with preservation of the open reading frame and with a probability score of > 0.3 were selected. The mapping uniqueness of each fusion contig was assessed using BLAT and the ones with ambiguous mapping were removed. After that, the supporting reads for each fusion event were visualized in IGV and remapped using BLAT. Similarly, only those correctly mapped to the annotated positions were counted as supporting reads. Finally, fusion transcripts with 3 or more supporting reads were selected for validation.
Fusion gene validation
Validations of fusion transcripts were carried out via PCR amplification of first strand cDNA followed by Illumina MiSeq sequencing of the amplicon pools. Validation primers (shown in Supplementary Table 11) were designed using Primer 3 program (see URLs). For each validation event, one oligonucleotide matching each end of the splitter was picked as forward and reverse primer to generate amplicons containing the breakpoints of the fusion RNAs. Briefly, 200 ng total RNA was used to prepared first strand cDNA and the first strand cDNA was amplified on Eppendorf Mastercycler ep384 (Eppendorf, Westbury, NY) with TaKaRa LA Taq DNA Polymerase, Hot-Start version (Cat no. RR042, Clontech). PCR products were examined and further purified. Amplicons with low yield were subjected to another round PCR program above with 2% of first round PCR products as template. Purified PCR products were equally pooled for Illumina library construction. 2 pools of 29 amplicons were sequenced on a Miseq sequencer.
Sequencing reads were aligned to human genome as described above for RNA-seq data. For each fusion event, its corresponding validation bam file was loaded to and visualized in IGV. The sequences of the supporting reads were extracted and remapped using BLAT to confirm the fusion breakpoints.
Mutation cross validation with RNA-seq data
RNA-seq data were also used to cross validate the somatic mutations identified by WES. For each somatic mutation reported by WES, SAMtools (v0.1.8)70 was used to count the read coverage for the variant and reference alleles in corresponding RNA-seq BAM files. A minimum of one high-quality supporting read was required to define a validated mutation. The validation results were summarized into the following 4 different conditions: validated with VAF > 5%; validated with VAF <5%; mutant allele not expressed if the mutation site was covered well but no mutant allele was detected; no information if no reads were mapped to the particular mutation site or gene.
TCR clonality analysis
The Gencode Genes (v18, see URLs) was used for annotation of the TCR loci. The igvtools (see URLs) was used to extract the base coverage of the corresponding TCR loci from RNA-seq BAM files for all samples. Wiggle files were generated for each sample and loaded into IGV for further analysis. The read scale was adjusted between the tumor and control samples for a better visualization.
SNP array and DNA copy number
Infinium SNP array assays were performed with Illumina Human OmniExpress12 v1-1 BeadChips (Cat. no. WG-312-1120). In brief, 200 ng genomic DNA was denatured, followed by isothermal whole genome amplification. The amplified DNA was enzymatically fragmented and hybridized to BeadChip for 16–24 hours at 48°C in hybridization oven. Twelve samples were processed in parallel on each BeadChip. A series of washing steps were conducted to remove unhybridized and non-specifically hybridized DNA fragments from the BeadChips, followed by allele-specific single-base extension reaction and multi-layer staining process. The coated BeadChips were imaged using Illumina iScan system. The SNP calls were collected using the Illumina GenomeStudio v2011.1 software. Cutoff for SNP call rate was set at 0.9 and data from samples passing this metric were used for further analyses.
For improved CNV analysis, B allele frequencies (BAF) were calculated and log2 R ratios (LRR) were extracted after re-clustering the raw data by applying the GenomeStudio clustering algorithms. The BAF and LRR data were then loaded into Nexus Copy Number v7.5 (BioDiscovery) to identify somatic DNA copy number alterations in SS patients. Systematic correction was applied to all samples with parameter settings refined for Illumina OmniExpress platform to correct the GC content related data waviness. The SNP-FASST2 segmentation algorithm was selected and matched paired analysis was performed following the manufacture's manual. Significant peaks were selected with a p-value cut-off of 0.05 and the aggregating frequency cut-off of 20%.
IL32 ELISA
The human CTCL cell lines HH, Hut78 and MJ were purchased from American Type Culture Collection (ATCC) (Manassas, VA). Other cell lines were kind gifts from Dr. Ivan Litvinov (Department of Medicine, McGill University, Canada). Cells were grown at 0.2–1.0×106/ml in HEPES-buffered RPMI 1640 medium as previously described14. Cell culture supernatants were collected on Day 3 after the media were refreshed. 8 ml of whole blood from patients were collected into a red-topped tube without additives, kept at RT for 30 minutes, and then centrifuged for 10 minutes at 1,000×g. The serum layers were then removed and aliquoted into small tubes and store at −80°C until use. The 5 normal human serum samples were purchased from different vendors and aliquot into small tubes and store at −80°C until use.
Soluble IL-32α in cell culture supernatants and human sera were measured using the Human IL-32α ELISA MAX™ Deluxe Sets (Biolegend, San Diego, CA). Briefly, the day before the assay, 100 μL/well of 1:200 capture antibody coating solution was added and the plate was incubated overnight at 4°C. Washing was performed immediately after each of the following steps to removed non-specific antibody binding. 200 μl/well of 1X Assay Diluent A was added and the plate was incubated at room temperature (RT) for 1 hour to block non-specific binding. After that, 100 μL/well of standards or samples were added and the plate was incubated at RT for 2 hours. The human IL-32α standard concentrations used were 500 pg/mL, 250 pg/mL, 125 pg/mL, 62.5 pg/mL, 31.3 pg/mL, 15.6 pg/mL, and 7.8 pg/mL, respectively. 1X Assay Diluent A serves as the zero standard (0 pg/mL). All standards were run in duplicate, and all samples were run in triplicate. After washing, 100 μL/well of 1:200 diluted Detection Antibody solution was added and the plate was incubated at RT for 1 hour. 100 μL/well of 1:1000 diluted Avidin-HRP solution was added and the plate was incubated at RT for 30 minutes. The plates were then developed with 100 μL/well of freshly mixed TMB Substrate Solution in the dark for 10-15 minutes followed by adding 100 μL/well of Stop Solution. The plates were read at 450 nm within 30 minutes. Concentrations were calculated and the data were shown as the mean of triplicate determinations.
IL32 Western Blot
The extracted cellular proteins were fractionated and transferred onto nitrocellulose membranes by electro-blotting. After being blocked in 5% milk in TBS supplemented with 0.05% Tween 20 for 1 hour at RT, the membranes were incubated with purified mouse anti-human IL-32αβδγ antibody IgG1, κ (Clone KU32-52, 1:1,000, Biolegend, San Diego, CA) overnight at 4 °C in 5% milk in TBST. The membranes were washed with TBST and then incubated with 1:1,000 HRP-conjugated goat anti-mouse polyclonal secondary antibodies (Cell Signaling, Beverly, MA) for 1 hour at RT. After washing with TBS, antibody binding was visualized using enhanced chemiluminescence (the SuperSignal™ West Pico Chemiluminescence Substrate, Thermo Fisher Scientific, Rockford, IL). The equivalent loading of proteins in each well was confirmed by Ponceau staining and actin control.
Supplementary Material
Table 1.
Significantly upregulated genes in SS patients that are important for T-cell function, pathogenesis or lineage significance
| Gene | Gene_ID | Expression in tumor | Expression in control | Fold change | p value | q value | Gene function |
|---|---|---|---|---|---|---|---|
| IL2RG | ENSG00000147168 | 250.79 | 41.81 | 6.0 | 4.90E-15 | 1.98E-10 | IL2, 4, 7 receptor activity, T-cell differentiation |
| IL32 | ENSG00000008517 | 869.92 | 31.76 | 27.4 | 7.59E-12 | 3.07E-07 | T-cell communication, proliferation |
| RAC2 | ENSG00000128340 | 269.14 | 50.12 | 5.4 | 2.49E-11 | 1.01E-06 | TCR signaling |
| LAT | ENSG00000213658 | 158.18 | 18.40 | 8.6 | 3.93E-11 | 1.59E-06 | TCR signaling |
| GIMAP4 | ENSG00000133574 | 206.62 | 7.29 | 28.3 | 5.19E-11 | 2.10E-06 | Lymphocyte apoptosis, Th1/Th2 differentiation |
| IKZF2 | ENSG00000030419 | 56.65 | 2.69 | 21.1 | 6.81E-11 | 2.75E-06 | T-reg lymphocyte development |
| CD3G | ENSG00000160654 | 117.53 | 16.90 | 7.0 | 4.08E-10 | 1.65E-05 | TCR signaling |
| TNFAIP8L2 | ENSG00000163154 | 13.38 | 0.61 | 22.1 | 6.35E-10 | 2.57E-05 | TCR function |
| SIT1 | ENSG00000137078 | 44.85 | 7.03 | 6.4 | 1.07E-09 | 4.32E-05 | TCR signaling |
| CD27 | ENSG00000139193 | 114.42 | 7.18 | 15.9 | 1.49E-08 | 6.02E-04 | T-cell differentiation, survival |
| PRKCQ | ENSG00000065675 | 32.50 | 6.45 | 5.0 | 3.43E-08 | 1.39E-03 | TCR signaling |
| CCR4 | ENSG00000183813 | 79.39 | 11.32 | 7.0 | 3.56E-07 | 1.44E-02 | T-cell migration into skin |
| CCR8 | ENSG00000179934 | 22.85 | 0.68 | 33.6 | 8.15E-07 | 3.30E-02 | T-reg lymphocyte survival |
| IKBKE | ENSG00000143466 | 19.06 | 2.57 | 7.4 | 1.57E-12 | 6.36E-08 | Oncogene: NF-κB and STAT signaling |
| CDK4 | ENSG00000135446 | 17.64 | 3.18 | 5.5 | 1.13E-11 | 4.56E-07 | Oncogene: cell cycle control |
| IL6R | ENSG00000160712 | 63.66 | 7.95 | 8.0 | 4.94E-10 | 2.00E-05 | IL6 receptor activity, immune response |
| HRAS | ENSG00000174775 | 13.27 | 2.59 | 5.1 | 5.59E-10 | 2.26E-05 | Oncogene |
| SLAMF6 | ENSG00000162739 | 63.68 | 2.41 | 26.4 | 7.56E-09 | 3.05E-04 | NK cell activation |
| TNFSF10 | ENSG00000121858 | 85.09 | 7.43 | 11.40 | 4.00E-08 | 1.61E-03 | Tumor necrosis factor, apoptosis |
| KLF8 | ENSG00000102349 | 11.02 | 0.41 | 26.8 | 1.15E-07 | 4.63E-03 | Cell cycle regulation, EMT |
| MYC | ENSG00000136997 | 41.45 | 8.13 | 5.1 | 3.83E-07 | 1.55E-02 | Oncogene: cell cycle, apoptosis |
TCR, T-cell receptor
ACKNOWLEDGEMENTS
This work was supported by research funding from the National Human Genome Research Institute (NHGRI, grant number: 5U54HG003273) and the Cancer Prevention Research Institute of Texas (CPRIT, grant number: RP121018) to D.W., the Drs. Martin and Dorothy Spatz Charitable Foundation (grant number: 00005840), the Blanche Bender Professorship in Cancer Research, and the MD Anderson Cancer Center Core Grant (grant number: CA16672) to M.D.. We thank Dr. Jianhong Hu, Dr. Mark Wang, Dr. Yi Han, Mr. Hsu Chao, Mr. Mark Gordon Evans for their excellent technical support. We thank Ms. Lee Sandra for help with sample intake, Ms. Joy Jayaseelan for project management. We thank Mr. Walker Hale, Ms. Divya Kalra, Ms. Shannon Dugan-Perez, and Ms. Jennifer Watt for their kind help with the data submission. Special thanks to Dr. Daniel Burgess and Dr. Michael Chase from Roche NimbleGen Inc. for their great help with the design and fast delivery of the custom capture array.
Footnotes
AUTHOR CONTRIBUTIONS
L.W. conducted the major bioinformatics analyses of all sequencing and SNP array data, integrated data from multiple platforms, wrote and revised the manuscript. X. N. contributed to the conduct of the research, data interpretation, and manuscript preparation. K.R.C. contributed to mutation signature analysis. B.Y., and J.S. contributed to the conduct of the research. X.Z. collected tumor specimens, performed DNA/RNA extraction and Western blot assay. L.X. and J.D contributed to the RNA-Seq and WES pipeline. Q.M. contributed to the fusion gene validation experiments. T.L. helped with the Serum and ELISA assay. D.M.M. and H.D. managed the production pipeline. L.A.D. contributed the somatic SMG analysis. R.A.G. contributed to the revision of the manuscript. M.D. recruited, consented, staged, characterized and cared for the patients, and supervised the skin biopsies. D.A.W. and M.D. conceived the study, supervised the research, and contributed to the writing and revision of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
URLs. dbGAP, http://www.ncbi.nlm.nih.gov/gap; MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp; IGV, http://www.broadinstitute.org/igv/; BLAT, https://genome.ucsc.edu/cgibin/hgBlat?command=start; Ensembl, http://www.ensembl.org/index.html; Gencode, http://www.gencodegenes.org; igvtools, http://www.broadinstitute.org/igv/igvtools; Primer 3, http://frodo.wi.mit.edu/primer3; Uniprot, http://www.uniprot.org.
Accession codes All sequencing and genotyping data have been deposited in the NCBI database of Genotypes and Phenotypes (dbGaP, see URLs) under accession phs000859.
References
- 1.Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood. 2010;116:767–71. doi: 10.1182/blood-2009-11-251926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girardi M, Heald PW, Wilson LD. The pathogenesis of mycosis fungoides. N Engl J Med. 2004;350:1978–88. doi: 10.1056/NEJMra032810. [DOI] [PubMed] [Google Scholar]
- 3.Kim YH, Liu HL, Mraz-Gernhard S, Varghese A, Hoppe RT. Long-term outcome of 525 patients with mycosis fungoides and Sezary syndrome: clinical prognostic factors and risk for disease progression. Arch Dermatol. 2003;139:857–66. doi: 10.1001/archderm.139.7.857. [DOI] [PubMed] [Google Scholar]
- 4.Olsen E, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood. 2007;110:1713–22. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 5.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. Journal of the American Academy of Dermatology. 2014;70:205, e1–16. doi: 10.1016/j.jaad.2013.07.049. quiz 221-222. [DOI] [PubMed] [Google Scholar]
- 6.Venkatarajan S, Duvic M. Sézary syndrome: an overview of current and future treatment options. Expert opinion on orphan drugs. 2014;2:3. [Google Scholar]
- 7.Herne KL, Talpur R, Breuer-McHam J, Champlin R, Duvic M. Cytomegalovirus seropositivity is significantly associated with mycosis fungoides and Sezary syndrome. Blood. 2003;101:2132–6. doi: 10.1182/blood-2002-07-2247. [DOI] [PubMed] [Google Scholar]
- 8.Talpur R, Bassett R, Duvic M. Prevalence and treatment of Staphylococcus aureus colonization in patients with mycosis fungoides and Sezary syndrome. Br J Dermatol. 2008;159:105–12. doi: 10.1111/j.1365-2133.2008.08612.x. [DOI] [PubMed] [Google Scholar]
- 9.Agar NS, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol. 2010;28:4730–9. doi: 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- 10.Scarisbrick JJ, et al. Prognostic factors, prognostic indices and staging in mycosis fungoides and Sezary syndrome: where are we now? Br J Dermatol. 2014;170:1226–36. doi: 10.1111/bjd.12909. [DOI] [PubMed] [Google Scholar]
- 11.Talpur R, et al. Long-term outcomes of 1,263 patients with mycosis fungoides and Sezary syndrome from 1982 to 2009. Clin Cancer Res. 2012;18:5051–60. doi: 10.1158/1078-0432.CCR-12-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidulich KA, Talpur R, Bassett RL, Duvic M. Overall survival in erythrodermic cutaneous T-cell lymphoma: an analysis of prognostic factors in a cohort of patients with erythrodermic cutaneous T-cell lymphoma. Int J Dermatol. 2009;48:243–52. doi: 10.1111/j.1365-4632.2009.03771.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim EJ, et al. Immunopathogenesis and therapy of cutaneous T cell lymphoma. J Clin Invest. 2005;115:798–812. doi: 10.1172/JCI24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni X, Zhang C, Talpur R, Duvic M. Resistance to activation-induced cell death and bystander cytotoxicity via the Fas/Fas ligand pathway are implicated in the pathogenesis of cutaneous T cell lymphomas. J Invest Dermatol. 2005;124:741–50. doi: 10.1111/j.0022-202X.2005.23657.x. [DOI] [PubMed] [Google Scholar]
- 15.Batista DA, et al. Multicolor fluorescence in situ hybridization (SKY) in mycosis fungoides and Sezary syndrome: search for recurrent chromosome abnormalities. Genes Chromosomes Cancer. 2006;45:383–91. doi: 10.1002/gcc.20302. [DOI] [PubMed] [Google Scholar]
- 16.Caprini E, et al. Identification of key regions and genes important in the pathogenesis of sezary syndrome by combining genomic and expression microarrays. Cancer Research. 2009;69:8438–8446. doi: 10.1158/0008-5472.CAN-09-2367. [DOI] [PubMed] [Google Scholar]
- 17.Mao X, Chaplin T, Young BD. Integrated genomic analysis of sézary syndrome. Genetics Research International. 2011;2011:980150. doi: 10.4061/2011/980150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao X, et al. Molecular cytogenetic characterization of Sezary syndrome. Genes Chromosomes Cancer. 2003;36:250–60. doi: 10.1002/gcc.10152. [DOI] [PubMed] [Google Scholar]
- 19.Vermeer MH, et al. Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Research. 2008;68:2689–2698. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 20.Braun FCM, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sezary syndrome. Leukemia. 2011;25:1494–1501. doi: 10.1038/leu.2011.101. [DOI] [PubMed] [Google Scholar]
- 21.Laharanne E, et al. CDKN2A-CDKN2B deletion defines an aggressive subset of cutaneous T-cell lymphoma. Mod Pathol. 2010;23:547–58. doi: 10.1038/modpathol.2009.196. [DOI] [PubMed] [Google Scholar]
- 22.Lamprecht B, et al. The tumour suppressor p53 is frequently nonfunctional in Sézary syndrome. The British Journal of Dermatology. 2012;167:240–246. doi: 10.1111/j.1365-2133.2012.10918.x. [DOI] [PubMed] [Google Scholar]
- 23.Vaqué JP, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014;123:2034–2043. doi: 10.1182/blood-2013-05-504308. [DOI] [PubMed] [Google Scholar]
- 24.Ungewickell A, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47:1056–60. doi: 10.1038/ng.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–9. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duvic M, Foss FM. Mycosis fungoides: pathophysiology and emerging therapies. Semin Oncol. 2007;34:S21–8. doi: 10.1053/j.seminoncol.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Horwitz SM. Novel therapies for cutaneous T-cell lymphomas. Clin Lymphoma Myeloma. 2008;8(Suppl 5):S187–92. doi: 10.3816/CLM.2008.s.015. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, et al. Novel somatic and germline mutations in intracranial germ cell tumours. Nature. 2014;511:241–5. doi: 10.1038/nature13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler DA, Wang L. From human genome to cancer genome: the first decade. Genome Res. 2013;23:1054–62. doi: 10.1101/gr.157602.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. 2015;7:269rv1. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu XS, Lonsdorf AS, Hwang ST. Cutaneous T-cell lymphoma: roles for chemokines and chemokine receptors. J Invest Dermatol. 2009;129:1115–9. doi: 10.1038/jid.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshie O, Matsushima K. CCR4 and its ligands: from bench to bedside. Int Immunol. 2015;27:11–20. doi: 10.1093/intimm/dxu079. [DOI] [PubMed] [Google Scholar]
- 35.Ni X, et al. Reduction of regulatory T cells by Mogamulizumab, a defucosylated anti-CC chemokine receptor 4 antibody, in patients with aggressive/refractory mycosis fungoides and Sezary syndrome. Clin Cancer Res. 2015;21:274–85. doi: 10.1158/1078-0432.CCR-14-0830. [DOI] [PubMed] [Google Scholar]
- 36.Duvic M, et al. Phase 1/2 study of mogamulizumab, a defucosylated anti-CCR4 antibody, in previously treated patients with cutaneous T-cell lymphoma. Blood. 2015 doi: 10.1182/blood-2014-09-600924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa M, et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J Exp Med. 2014;211:2497–2505. doi: 10.1084/jem.20140987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumoto R, et al. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Hara H, et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–75. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, et al. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nature Immunology. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 41.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science (New York, N.Y.) 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 43.Dereure O, Portales P, Clot J, Guilhou JJ. Decreased expression of Fas (APO-1/CD95) on peripheral blood CD4+ T lymphocytes in cutaneous T-cell lymphomas. Br J Dermatol. 2000;143:1205–10. doi: 10.1046/j.1365-2133.2000.03889.x. [DOI] [PubMed] [Google Scholar]
- 44.Jones CL, et al. Downregulation of Fas gene expression in Sézary syndrome is associated with promoter hypermethylation. The Journal of Investigative Dermatology. 2010;130:1116–1125. doi: 10.1038/jid.2009.301. [DOI] [PubMed] [Google Scholar]
- 45.Postigo AA, Dean DC. Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol Cell Biol. 1999;19:7961–71. doi: 10.1128/mcb.19.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higashi Y, et al. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–79. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakahata S, Yamazaki S, Nakauchi H, Morishita K. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-beta1-mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene. 2010;29:4157–69. doi: 10.1038/onc.2010.172. [DOI] [PubMed] [Google Scholar]
- 48.Hidaka T, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112:383–93. doi: 10.1182/blood-2008-01-131185. [DOI] [PubMed] [Google Scholar]
- 49.Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–62. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagl NG, Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nam HJ, et al. The ERK-RSK1 activation by growth factors at G2 phase delays cell cycle progression and reduces mitotic aberrations. Cell Signal. 2008;20:1349–58. doi: 10.1016/j.cellsig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol. 2013;14:1212–8. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- 54.Suga H, et al. The role of IL-32 in cutaneous T-cell lymphoma. The Journal of Investigative Dermatology. 2014;134:1428–1435. doi: 10.1038/jid.2013.488. [DOI] [PubMed] [Google Scholar]
- 55.Ohmatsu H, et al. IL32 Is Progressively Expressed in Mycosis Fungoides Independent of Helper T-cell 2 and Helper T-cell 9 Polarization. Cancer Immunology Research. 2014;2:890–900. doi: 10.1158/2326-6066.CIR-13-0199-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim KH, et al. Interleukin-32 monoclonal antibodies for immunohistochemistry, Western blotting, and ELISA. J Immunol Methods. 2008;333:38–50. doi: 10.1016/j.jim.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 57.Huether R, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vega F, et al. Clonal heterogeneity in mycosis fungoides and its relationship to clinical course. Blood. 2002;100:3369–73. doi: 10.1182/blood.V100.9.3369. [DOI] [PubMed] [Google Scholar]
- 59.Jackow CM, et al. Association of erythrodermic cutaneous T-cell lymphoma, superantigen-positive Staphylococcus aureus, and oligoclonal T-cell receptor V beta gene expansion. Blood. 1997;89:32–40. [PubMed] [Google Scholar]
- 60.Tan RS, Butterworth CM, McLaughlin H, Malka S, Samman PD. Mycosis fungoides--a disease of antigen persistence. Br J Dermatol. 1974;91:607–16. doi: 10.1111/j.1365-2133.1974.tb12449.x. [DOI] [PubMed] [Google Scholar]
- 61.Spits H. Development of alphabeta T cells in the human thymus. Nat Rev Immunol. 2002;2:760–72. doi: 10.1038/nri913. [DOI] [PubMed] [Google Scholar]
- 62.Murphy KP, Mowat A, Weaver CT. Janeway's Immunobiology. Garland Science; 2011. [Google Scholar]
- 63.Bradford PT, Devesa SS, Anderson WF, Toro JR. Cutaneous lymphoma incidence patterns in the United States: a population-based study of 3884 cases. Blood. 2009;113:5064–73. doi: 10.1182/blood-2008-10-184168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takashima A. Establishment of Fibroblast Cultures. in Current Protocols in Cell Biology. John Wiley & Sons, Inc.; 2001. [DOI] [PubMed] [Google Scholar]
- 65.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–2. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–9. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 69.McPherson A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.