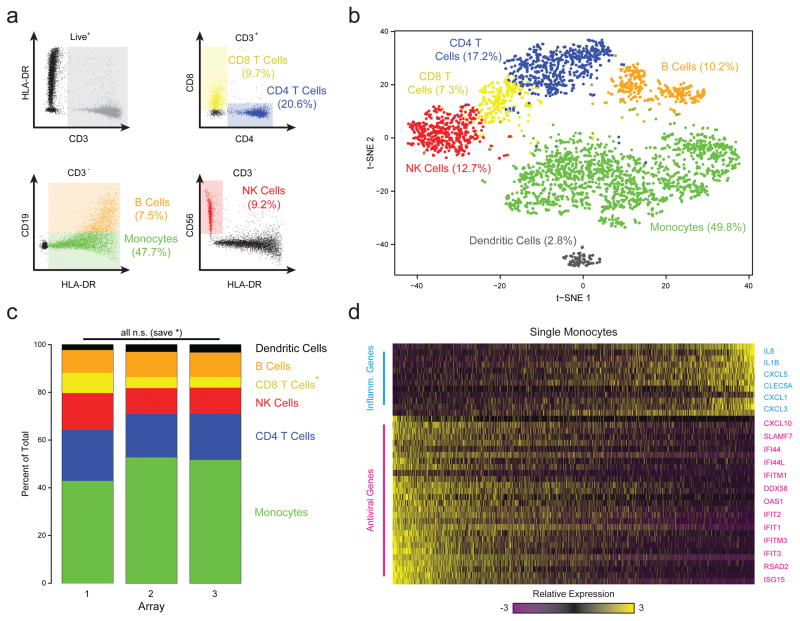
Figure 2. Combined Image Cytometry and scRNA-Seq of Human PBMCs.
(a) The hierarchical gating scheme (with the frequencies of major cell subpopulations) used to analyze PBMCs that had been labeled with a panel of fluorescent antibodies, loaded onto three replicate arrays and imaged prior to bead loading and transcript capture (Methods). Myeloid cells (green) were identified as the population of hCD3(-) HLA-DR(+) CD19(-) cells; B cells (orange) as the subset of hCD3(-) HLA-DR(+) CD19 (+) cells; CD4 T cells (blue) as the subset of CD3(+) CD4(+) cells; CD8 T cells (yellow) as the CD3(+) CD8(+) subset of cells; and, NK cells (red) as the subset of CD3(-) HLA-DR (-) CD56 (+) CD16(+) cells. (b) t-SNE visualization of single-cell clusters identified among 3,694 human Seq-Well PBMCs single-cell transcriptomes recovered from the imaged array and the two additional ones (Methods; Supplementary Fig. 10–12). Clusters (subpopulations) are labeled based on annotated marker gene (Supplementary Fig. 10). (c) The distribution of transcriptomes captured on each of the 3 biological replicate arrays, run on separate fractions of the same set of PBMCs. All shifts are insignificant save for a slightly elevated fraction of CD8 T cells in array 1 (*, p=1.0×10−11; Chi-square Test, Bonferroni-corrected). (d) A heatmap showing the relative expression level of a set of inflammatory and antiviral genes among cells identified as monocytes.