Abstract
Single-cell RNA-Seq can precisely resolve cellular states but application to sparse samples is challenging. Here, we present Seq-Well, a portable, low-cost platform for massively-parallel single-cell RNA-Seq. Barcoded mRNA capture beads and single cells are sealed in an array of subnanoliter wells using a semi-permeable membrane, enabling efficient cell lysis and transcript capture. We characterize Seq-Well using species-mixing experiments and PBMCs, and profile thousands of primary human macrophages exposed to tuberculosis.
MAIN
The emergence of single-cell genomics has empowered new strategies for identifying the cellular and subcellular drivers of biological phenomena1–19. Patterns in genome-wide mRNA expression measured by single-cell RNA-Seq (scRNA-Seq) can be leveraged to uncover distinct cell types, states and circuits within cell populations and tissues1–5,9–13. The unprecedented view of cellular phenotypes scRNA-Seq affords could help transform our understanding of healthy and diseased behaviors, and guide the rational selection of precision diagnostics and therapies, if it could be broadly and easily applied to low-input (≤104 cells) clinical specimens.
Typically, scRNA-Seq has involved isolating and lysing individual cells, then independently reverse transcribing and amplifying their mRNA before generating barcoded libraries that are pooled for sequencing. Although manual picking2,5,8, FACS-sorting1,3,4 or integrated microfluidic circuits7,9,10 can isolate single cells, one-cell-one-sample approaches are constrained fundamentally in scale by costs, time, and labor. Recently, massively-parallel methods have emerged that assign unique barcodes to each cell’s mRNAs during reverse transcription, enabling ensemble processing while retaining single-cell resolution. These methods typically yield single-cell libraries of lower complexity, but higher throughput reduces the impact of the technical and intrinsic noise associated with each cell in analyses11,12. The most common variant is microfluidic devices that generate reverse-emulsion droplets to serially couple single cells with uniquely-barcoded mRNA capture beads11,12. Droplet-based techniques, however, can have inefficiencies in encapsulation, introduce technical noise through differences in cell lysis time, and require specialized equipment, limiting where, when, and with what scale scRNA-Seq can be performed.
One alternative is to use arrays of subnanoliter wells loaded by gravity. Operational simplicity reduces the need for peripheral equipment, decreases dead volumes, and facilitates parallelization. As proof-of-principle, cells and beads have been co-confined in unsealed nanowell arrays to perform targeted single-cell transcriptional profiling13, yet the use of an open-array format significantly limits capture efficiency and increases cross-contamination (Supplementary Fig. 1). To avoid these issues, nanowells have also been combined with microfluidic channels that facilitate oil-based single-cell isolation via fluid exchange14. Nevertheless, this design limits buffer exchange and necessitated integrated temperature and pressure controllers, impacting ease-of-use and portability15. Semi-porous-membrane-covered nanowells have been used to link pairs of specific transcripts from single cells16; however, transcript capture and sealing efficiency were not addressed, and unique single-cell libraries were not achieved using many beads per well.
To overcome these assorted challenges, we have developed Seq-Well, a portable, simple platform for massively-parallel scRNA-Seq (Supplementary Fig. 2). Similar to previous nanowell-based implementations, Seq-Well confines single cells and barcoded poly(dT) mRNA capture beads in a PDMS array of ~86,000 subnanoliter wells. Designing well dimensions to accommodate only one bead enables single-bead loading efficiencies of ~95% (Figure 1a, Supplementary Fig. 3a; Supplementary Video 1). A simplified cell-loading scheme, in turn, enables capture efficiencies around 80% (Methods; Supplementary Fig. 3b), with a rate of dual occupancy that can be tuned by adjusting the number of cells loaded and visualized prior to processing (Supplementary Fig. 3c).
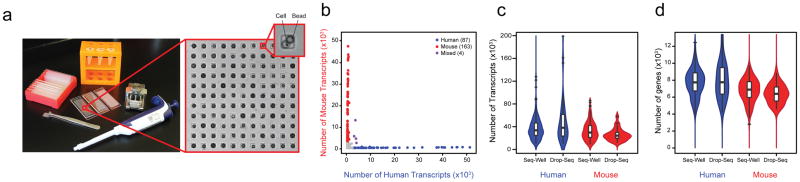
Figure 1. Seq-Well: A Portable, Low-Cost Platform for High-Throughput Single-Cell RNA-Seq of Low-Input Samples.
(a) Photograph of equipment and array used to capture and lyse cells, respectively. (b) Transcripts captured from a mix of human (HEK293) and mouse (NIH/3T3) cells reveal distinct transcript mapping and single-cell resolution. Human (mouse) cells (> 2,000 human (mouse) transcripts and < 1,000 mouse (human) transcripts) are shown in blue (red). Among the 254 cells identified, 1.6% (shown in purple) had a mixed phenotype. (c,d) Violin plots of the number of transcripts (c) and genes (d) detected in human or mouse single-cell libraries generated by Seq-Well or Drop-Seq (Ref. 12; Center-line: Median; Limits: 1st and 3rd Quartile; Whiskers: +/− 1.5 IQR; Points: Values > 1.5 IQR). Using Seq-Well (Drop-Seq), an average of 37,878 (48,543) transcripts or 6,927 (7,175) genes were detected among human HEK cells (n = 159 for Seq-Well; n = 48 for Drop-Seq) and an average of 33,586 (26,700) transcripts or 6,113 (5,753) genes were detected among mouse 3T3 cells (n = 172 for Seq-Well; n = 27 for Drop-Seq) at an average read depth of 164,238 (797,915) reads per human HEK cell and an average read depth of 152,488 (345,117) read per mouse 3T3 cell.
Importantly, Seq-Well uniquely leverages selective chemical functionalization to facilitate reversible attachment of a semi-permeable polycarbonate membrane (10 nm pore size) in physiologic buffers. This trait enables rapid solution exchange for efficient cell lysis but traps biological macromolecules, increasing transcript capture during hybridization and reducing cross-contamination (Supplementary Fig. 4a; Supplementary Protocol; Supplementary Video 2). The array’s unique three-layer surface functionalization comprises an amino-silane base20 crosslinked to bifunctional poly(glutamate)/chitosan top via a p-Phenylene diisothiocyante intermediate (Methods; Supplementary Fig. 4); this bifunctional top, with poly(glutamate) coating the inner surfaces of the nanowells (where cells are lysed) and chitosan the array’s top surface (where the membrane binds), prevents non-specific binding of RNA to the array and efficient sealing, respectively (Methods; Supplementary Protocol; Supplementary Fig. 4b,c). To test sealing and buffer exchange, we monitored the fluorescence of dye-labeled, cell-bound antibodies before and after adding a guanidinium-based lysis buffer. We observed rapid diffusion of the antibodies throughout the wells within five minutes of buffer addition and, unlike unsealed or previously-described, membrane-covered BSA-blocked arrays16, little change in fluorescent signal over 30 minutes, suggesting robust retention of biological macromolecules despite use of a strong chaotrope (Methods; Supplementary Fig. 5).
After lysis, cellular mRNAs are captured by bead-bound poly(dT) oligonucleotides that also contain a universal primer sequence, a cell barcode, and a unique molecular identifier (UMI) (Methods; Supplementary Table 1). Next, the membrane is peeled off and the beads are removed for subsequent bulk reverse transcription, amplification, library preparation and paired-end sequencing, as previously described12 (Methods). Critically, beyond a disposable array and membrane, Seq-Well only requires a pipette, a manual clamp, an oven, and a tube rotator to achieve stable, barcoded single-cell cDNAs (Fig. 1a), enabling it to be performed almost anywhere.
To assess transcript capture efficiency and single-cell resolution, we profiled a mixture of 5×103 human (HEK293) and 5×103 mouse (3T3) cells using Seq-Well. The average fraction of reads mapping to exonic regions was 77.5% (Supplementary Fig. 6), demonstrating high quality libraries. Shallow sequencing from a fraction of an array revealed highly organism-specific libraries, suggesting single-cell resolution and minimal cross-contamination (Fig. 1b; Supplementary Fig. 7a–c). In the absence of membrane sealing, by comparison, we obtained poor transcript and gene detection, and substantial cross-contamination (Supplementary Fig. 1). From deeper sequencing of a fraction of a second array, we detected an average of 37,878 mRNA transcripts from 6,927 genes in HEK cells and 33,586 mRNA transcripts from 6,113 genes in 3T3 cells, comparable to a droplet-based approach using the same mRNA capture beads (Drop-Seq)12 (Fig. 1c,d & Supplementary Fig. 7&8). Upon matched-read downsampling, we also observed levels of transcript and gene detection consistent with other massively-parallel bead-based scRNA-Seq methods (Methods; Supplementary Fig. 7d–g). Moreover, we found strong correlations between bulk RNA-seq data and populations constructed in silico from individual HEK cells (R=0.751±0.073–0.983±0.0001 for populations of 1–1,000 single cells, respectively), suggesting representative cell and transcript sampling (Methods; Supplementary Fig. 9).
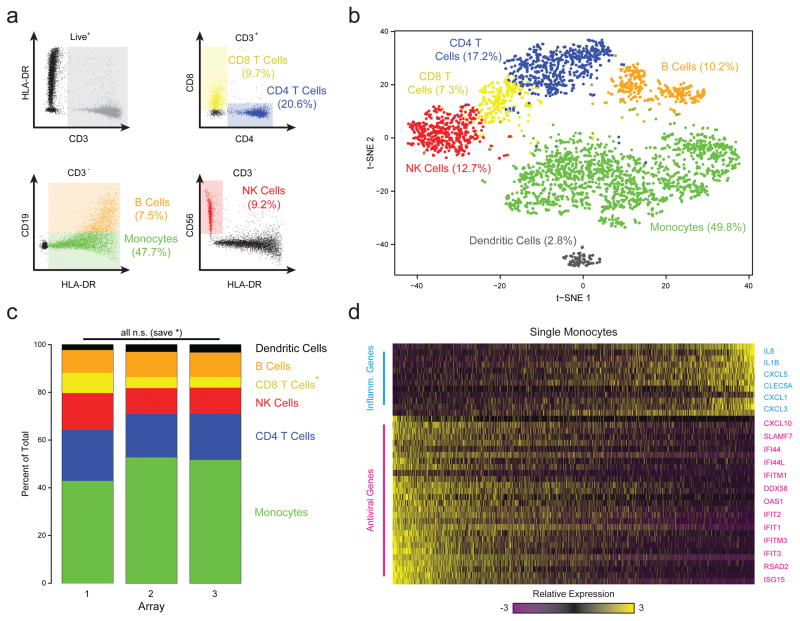
Next, to examine the ability of Seq-Well to resolve populations of cells in complex primary samples, we loaded human peripheral blood mononuclear cells (PBMCs) into arrays in triplicate prior to beads, allowing us to perform on-array multi-color imaging cytometry (Methods; Fig. 2a,b, Supplementary Tables 2&3). Sequencing one-third of the beads recovered from each array yielded 3,694 high-quality single-cell libraries (Methods). Unsupervised graph-based clustering revealed unique subpopulations corresponding to major PBMC cell types (Methods; Fig. 2b, Supplementary Fig. 10–12; Supplementary Table 4). Each array yielded similar subpopulation frequencies (Fig. 2c), with detection efficiencies comparable to other massively-parallel technologies (Supplementary Fig. 13). The proportion of each subpopulation determined by sequencing also matched on-array immunophenotyping results (Fig. 2a,b). Critically, sequencing provides additional information: in addition to resolving dendritic cells from monocytes (Fig. 2b), we found significant variation among the monocytes (captured in PC3) due to differential expression of inflammatory and anti-viral gene programs (Fig. 2d)1,3. Overall, characterizing a sample in two ways using a single platform increases the amount of the information that can be extracted from a precious specimen, while also allowing analysis of one measurement in light of the other.
Figure 2. Combined Image Cytometry and scRNA-Seq of Human PBMCs.
(a) The hierarchical gating scheme (with the frequencies of major cell subpopulations) used to analyze PBMCs that had been labeled with a panel of fluorescent antibodies, loaded onto three replicate arrays and imaged prior to bead loading and transcript capture (Methods). Myeloid cells (green) were identified as the population of hCD3(-) HLA-DR(+) CD19(-) cells; B cells (orange) as the subset of hCD3(-) HLA-DR(+) CD19 (+) cells; CD4 T cells (blue) as the subset of CD3(+) CD4(+) cells; CD8 T cells (yellow) as the CD3(+) CD8(+) subset of cells; and, NK cells (red) as the subset of CD3(-) HLA-DR (-) CD56 (+) CD16(+) cells. (b) t-SNE visualization of single-cell clusters identified among 3,694 human Seq-Well PBMCs single-cell transcriptomes recovered from the imaged array and the two additional ones (Methods; Supplementary Fig. 10–12). Clusters (subpopulations) are labeled based on annotated marker gene (Supplementary Fig. 10). (c) The distribution of transcriptomes captured on each of the 3 biological replicate arrays, run on separate fractions of the same set of PBMCs. All shifts are insignificant save for a slightly elevated fraction of CD8 T cells in array 1 (*, p=1.0×10−11; Chi-square Test, Bonferroni-corrected). (d) A heatmap showing the relative expression level of a set of inflammatory and antiviral genes among cells identified as monocytes.
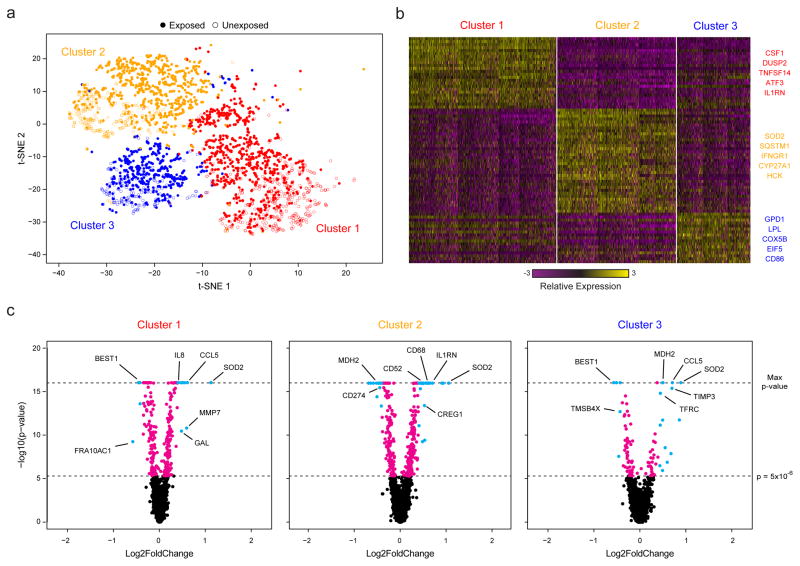
Finally, to test the portability of Seq-Well, we profiled primary human macrophages exposed to M. tuberculosis (H37Rv) in a BSL3 facility (Methods). In total, we recovered 14,218 (of 40,000 possible) macrophages with greater than 1,000 mapped transcripts from a TB-exposed and an unexposed array. Unsupervised analysis of 4,638 cells with greater than 5,000 transcripts per cell revealed five distinct clusters (Fig. 3a,b & Supplementary Fig. 14a,b; Supplementary Table 5). Two had lower transcript capture and high mitochondrial gene expression (suggestive of low quality libraries)17, and were removed; the remaining three (2,560 cells) were identified in both the exposed and unexposed samples (Fig. 3a, Supplementary Fig. 14c,d&15), and likely represent distinct sub-phenotypes present in the initial culture.
Figure 3. Sequencing of TB-Exposed Macrophages in a BSL3 Facility Using Seq-Well.
(a) t-SNE visualization of single-cell clusters identified among 2,560 macrophages (1,686 exposed, solid circles; 874 unexposed, open circles) generated using 5 principal components across 377 variable genes (Methods). (b) Marker genes for the 3 phenotypic clusters of macrophages highlighted in (a). (c) Volcano plots of differential expression between exposed and unexposed macrophages within each cluster showing genes enriched in cells exposed to M. tuberculosis. In each plot, a p-value threshold of 5.0 ×10−16 based on a likelihood ratio test was used to establish statistical significance, while a log2-fold change threshold of 0.4 was used to determine differential expression. Genes with p-values less than 5.0×10−6 are shown in cyan and absolute log2-fold changes greater than 0.4; In magenta are genes with p-values less than 5.0×10−6 but absolute log2-fold changes less than 0.4; and, in black, are genes with p-values greater than 5.0×10−6 and absolute log2-fold changes less than 0.4.
We next examined common and cluster-specific gene enrichments (Methods). Although Clusters 1 and 3 did not present strong stimulation-independent enrichments, Cluster 2 uniquely expressed several genes associated with metabolism (Supplementary Tables 6&7). Intriguingly, within each cluster, we observed pronounced shifts in gene expression in response to M. tuberculosis (Methods; Fig. 3c & Supplementary Table 8), with common enrichments for gene sets previously observed in response to intracellular infection, LPS stimulation, and activation of TLR7/8 (Supplementary Tables 9&10). Cluster 1 uniquely displayed stimulation-induced shifts in several genes associated with cell growth, Cluster 3 in transcripts associated with hypoxia, and Cluster 2, again, in genes linked to metabolism. Overall, these data suggest that basal cellular heterogeneity may influence ensemble tuberculosis responses. Equally importantly, they demonstrate the ability of Seq-Well to acquire large numbers of single-cell transcriptomes in challenging experimental environments.
In conclusion, Seq-Well is a robust platform for scalable, single-cell transcriptomics applicable to almost any cellular suspension for which a reference genome or transcriptome exists. The technique is inexpensive, user-friendly, portable, and efficient, enabling scRNA-Seq to accelerate scientific and clinical discovery, even when working with limited samples. Furthermore, the ability to measure protein secretion and cell surface expression on the same platform18,19 foreshadows multi-omic single-cell measurements at scale. As such, our platform may prove to be a potent tool for empowering a new era of precision science and medicine.
METHODS
Methods and any associated references are available in the online version of the paper.
Supplementary Material
Supplementary Table 1 | Oligo Sequences. Sequences of oligos used in Seq-Well. (1) The barcoded bead sequence is constructed on the surface of the bead, and cell barcodes are generated through split and pool synthesis. (2) The template switching oligo (TSO) is used to tag the 5′ end of captured mRNA using a reverse transcriptase enzyme with terminal transferase activity. (3) Sequence for PCR primer used to perform whole-transcriptome amplification (WTA) PCR reaction following reverse transcription and ExoI digestion. (4) Sequence that selectively primes the bead-specific SMART sequence during the post-tagmentation step-out PCR, which appends a P5 sequencing adapter. (5) Primer used during sequencing that selectively primes the bead-specific primer site to initiate sequencing of the barcode and UMI in Illumina Read 1.
Supplementary Table 2 | Microscope Settings. The excitation light wavelengths, emission filters, exposure times, light source intensity and camera gain settings used to capture the fluorescence of the indicated fluorophore are displayed.
Supplementary Table 3 | Gene Expression Matrix for PBMCs. UMI count matrix for the 4,296 PBMCs, labeled by array, that had at least 10,000 reads, 1,000 transcripts, and 500 genes, with at least 65% bases mapping to the transcriptome.
Supplementary Table 4 | PBMC Cluster Enrichments. Lists of genes enriched within each PBMC cluster (B cells, CD4 T Cells, CD8 T cells, Dendritic cells, Monocytes, NK Cells) based on a likelihood-ratio test in which members of each cluster are compared to members of all other clusters.
Supplementary Table 5 | Gene Expression Matrix for Mtb-Exposed Monocyte-Derived Macrophages and Unexposed Control Cells. UMI count matrix for the 4,638 monocyte-derived macrophages, labeled by exposure, that had at least 5,000 mapped transcripts.
Supplementary Table 6 | TB Cluster Enrichments. We examined sets of enriched sets of genes among all cells (irrespective of TB exposure) within Cluster 1, 2 and 3 using the find.markers function in Seurat, which implements a ‘roc’ test to identify relative expression differences. (a) From this analysis, we identified sets of genes exclusively enriched in each cluster but not the others. For each of the identified gene sets, we performed gene set enrichment analysis in DAVID and GSEA. For analysis in DAVID, we compared each gene list for enrichment among GO terms and curated pathways against a background list of 9045 genes contained in the DAVID database that were detected in at least 5% of cells. For analysis using GSEA, we compared each gene list to the complete database of gene sets contained within GSEA. (b, c) Within Cluster 1, we observe unique enrichment of 134 genes related to TNF-alpha signaling, inflammation, immune response and LPS response among 1099 cells. (d, e) In Cluster 2, we observe exclusive enrichment of 251 genes among 904 cells that distinguish monocytes from dendritic cells in culture and characterize TNF-alpha signaling. (f, g) Finally, in Cluster 3, we observe unique enrichment of 118 genes among 557 cells related to specifically to hypoxia, LPS stimulation, TNF-alpha signaling and apoptosis.
Supplementary Table 7 | Cluster Enrichments between Exposure Groups. Initially, we separately identified enriched genes among exposed and unexposed cells within Clusters 1, 2 and 3 using the find.markers function in Seurat. In total, we identified 18 genes with conserved enrichment among TB exposed cells within Clusters 1, 2 and 3. For each cluster, we identified the set of genes enriched among exposed cells and unexposed cells within each cluster. (a) We identified 28 enriched genes that were conserved among exposed cells across clusters and 31 conserved genes among unexposed, of which 18 were conserved between exposed and unexposed. We identified 38 genes uniquely enriched among exposed cells in Cluster 1 and 54 genes uniquely enriched among unexposed cells, of which 5 were conserved between exposed and unexposed cells within Cluster 1. We identified 134 genes unique to Cluster 2 among exposed cells and 200 genes unique to Cluster 2 among unexposed cells, of which 35 were conserved between exposed and unexposed cells within Cluster 2. In Cluster 3, we identified 43 genes unique to cluster 3 among exposed cells and 44 genes unique to Cluster 3 among unexposed cells, of which 9 were conserved between exposed and unexposed cells within Cluster 3. We performed gene set enrichment analyses for single gene list using DAVID and GSEA. In DAVID, we specified a background list of 9045 genes and examined enrichments within GO terms and curated pathways. For the analysis in GSEA, we made comparisons of gene lists to the complete database of gene sets within GSEA. (b, c) Among the 18 genes conserved across clusters in both exposed and unexposed cells, we observed strong enrichment for LPS response, TNF-alpha signaling, phagosome formation and macrophage activation. (d, e) Among the 5 genes unique to Cluster 1 conserved between exposed and unexposed cells, we observed enrichment of PI3K-Akt signaling and immune activation. (f, g) Among the 35 genes unique to Cluster 2 conserved between exposed and unexposed cells, we observed enrichment of genes related to monocyte culture and the coagulation cascade. (h, i) Among the 9 genes unique to Cluster 3 conserved between exposed and unexposed, we observed enrichment of genes up-regulated by HGF and apoptosis.
Supplementary Table 8 | Differentially Expressed Genes between TB Exposed and Unexposed Cells within Each Cluster. We performed a likelihood ratio test to identify genes differentially expressed between TB exposed and unexposed cells within each cluster. (a) Differential expression results between 673 TB-exposed and 426 unexposed cells in Cluster 1. (b) Differential expression results between 627 TB-exposed and 277 unexposed cells in Cluster 2. (c) Differential expression results between 386 TB-exposed and 171 unexposed cells in Cluster 3.
Supplementary Table 9 | TB Infection by Cluster Enrichments. Initially, we performed a LRT within each cluster to identify genes differentially expressed between TB exposed and unexposed cells. For each cluster, we created lists of genes differentially expressed with p-values less than 5.0×10−6 within each cluster (Figure 3c). (a) We then compared these lists to identify genes that are differentially expressed genes exclusively within each cluster. We also identified a list of 37 genes detected as differentially expressed across all clusters. We then performed gene set enrichment analysis in DAVID and GSEA to examine functional enrichment of the identified gene sets (i.e. Genes conserved across and unique to each cluster). We performed analysis in DAVID, comparing genes 37 conserved genes, 22 genes unique to Cluster 1, 142 genes unique to Cluster 2, and 40 genes unique to Cluster 3 to a background list of 9,381 genes expressed in at least 5% of filtered cells (Methods). Using GSEA, we made comparisons of the above gene list to the complete list of curated gene sets within the GSEA database (MSigDB v5.1: http://software.broadinstitute.org/gsea/msigdb/index.jsp). (b,c) Within Cluster 1, we observed unique enrichment of genes related to growth, proliferation and cell cycle. (d, e) Within Cluster 2, we observed enrichment of genes that identify monocyte and dendritic cell culture in addition to proliferation. (f, g) Within Cluster 3, we observed unique enrichment of genes related to hypoxia, oxidative stress and oxygen homeostasis.
Supplementary Table 10 | GSEA Comparisons of Exposed and Unexposed Cells within Each Cluster. We performed comparisons between M. tuberculosis exposed and unexposed cells within each cluster using GSEA. For each cluster we created .gct files containing normalized expression data for every cells within each cluster and assigned phenotypes (i.e. TB exposed vs. unexposed) to each cell using a .cls file. We then performed gene set enrichment analysis for each cluster across the complete gene set database in GSEA with 1000 permutations of assigned phenotype. (a,b) In cluster 1, we observed enrichment of dendritic cell maturation, monocytes in culture, response to L. donovani, and TNF-alpha signaling among 673 TB exposed cells and relative enrichment of ribosomal genes and protein synthesis among 426 unexposed cells. (c,d) In Cluster 2, we observed enrichment of LPS response, dendritic cell maturation, IL1 stimulation and response to TGF-beta among 627 TB exposed cells and relative enrichment of housekeeping functions, ribosomal genes and translation among 277 TB unexposed cells. (e,f) In Cluster 3, we observed enrichment of among delayed response to LPS (48 response), TLR7/8 stimulation, inflammatory response, intracellular infection and TNF signaling among 386 TB exposed cells and relative enrichment of housekeeping functions (ribosome, translation, actin) among 171 unexposed cells. (g,h) In Cluster 4, we observed enrichment of mitochondrial gene signatures, oxidative phosphorylation, hypoxia response and interferon response among 74 TB exposed cells and enrichment of ribosomal genes and translation among 975 unexposed cells. (i,j) In Cluster 5, we observed enrichment of LPS stimulation, TNF signaling, sepsis and dendritic cells maturation among 988 TB exposed cells and enrichment of ribosomal proteins and translation among 41 unexposed cells.
Acknowledgments
We thank K. Shekar, T. Tickle, and M. Xie for fruitful discussions.
This work was supported by the Searle Scholars Program (AKS), the Beckman Young Investigator Program (AKS), a NIH New Innovator Award DP2 OD020839 (AKS), U24 AI11862-01 (AKS), P50 HG006193 (AKS), the Bill and Melinda Gates Foundation grant 03629000189 (AKS, JCL, SF), the Ragon Institute (AKS, SF), the Burroughs Wellcome Foundation (SF), P30 AI060354 (SF), DP3 DK09768101 (JCL), P01 AI045757 (JCL), R21 AI106025 (JCL), R56 AI104274 (JCL), the W.M. Keck Foundation (JCL), and the U. S. Army Research Office through the Institute for Soldier Nanotechnologies, under contract number W911NF-13-D-0001 (JCL). This work was also supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. JCL is a Camille Dreyfus Teacher-Scholar.
Footnotes
DATA AVAILABILITY.
All RNA-Seq data are available in GEO under Accession Number GSE92495.
AUTHOR CONTRIBUTIONS.
TMG, MHW, TKH, JCL, and AKS developed the concepts and designed the study. TG, MHW, TKH, and BDB performed the experiments. All authors analyzed and interpreted the data. TMG, MHW, TKH, JCL, and AKS wrote the manuscript with feedback from all authors.
COMPETING FINANCIAL INTERESTS.
T.M. Gierahn, M.H. Wadsworth II, T.K. Hughes, J.C. Love, A.K. Shalek, and Institutions The Broad Institute and the Massachusetts Institute of Technology have filed a patent application that relates to Seq-Well, compositions of matter, the outlined experimental and computational methods, and uses thereof.
References
- 1.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lohr JG, et al. Whole exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nature biotechnology. 2014;32:479. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shalek AK, et al. Single cell RNA Seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nature methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 10.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan HC, Fu GK, Fodor SP. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 14.Bose S, et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome biology. 2015;16:1. doi: 10.1186/s13059-015-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J, Sims PA. An Automated Microwell Platform for Large-Scale Single Cell RNA-Seq. Scientific Reports. 2016;6 doi: 10.1038/srep33883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeKosky BJ, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nature biotechnology. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilicic T, et al. Classification of low quality cells from single-cell RNA-seq data. Genome biology. 2016;17:1. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka YJ, et al. Single-cell analysis of the dynamics and functional outcomes of interactions between human natural killer cells and target cells. Integrative Biology. 2012;4:1175–1184. doi: 10.1039/c2ib20167d. [DOI] [PubMed] [Google Scholar]
- 19.Han Q, et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proceedings of the National Academy of Sciences. 2012;109:1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinberg G, Stromsborg K, Thomas L, Barker D, Zhao C. Strategies for covalent attachment of DNA to beads. Biopolymers. 2004;73:597–605. doi: 10.1002/bip.20006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 | Oligo Sequences. Sequences of oligos used in Seq-Well. (1) The barcoded bead sequence is constructed on the surface of the bead, and cell barcodes are generated through split and pool synthesis. (2) The template switching oligo (TSO) is used to tag the 5′ end of captured mRNA using a reverse transcriptase enzyme with terminal transferase activity. (3) Sequence for PCR primer used to perform whole-transcriptome amplification (WTA) PCR reaction following reverse transcription and ExoI digestion. (4) Sequence that selectively primes the bead-specific SMART sequence during the post-tagmentation step-out PCR, which appends a P5 sequencing adapter. (5) Primer used during sequencing that selectively primes the bead-specific primer site to initiate sequencing of the barcode and UMI in Illumina Read 1.
Supplementary Table 2 | Microscope Settings. The excitation light wavelengths, emission filters, exposure times, light source intensity and camera gain settings used to capture the fluorescence of the indicated fluorophore are displayed.
Supplementary Table 3 | Gene Expression Matrix for PBMCs. UMI count matrix for the 4,296 PBMCs, labeled by array, that had at least 10,000 reads, 1,000 transcripts, and 500 genes, with at least 65% bases mapping to the transcriptome.
Supplementary Table 4 | PBMC Cluster Enrichments. Lists of genes enriched within each PBMC cluster (B cells, CD4 T Cells, CD8 T cells, Dendritic cells, Monocytes, NK Cells) based on a likelihood-ratio test in which members of each cluster are compared to members of all other clusters.
Supplementary Table 5 | Gene Expression Matrix for Mtb-Exposed Monocyte-Derived Macrophages and Unexposed Control Cells. UMI count matrix for the 4,638 monocyte-derived macrophages, labeled by exposure, that had at least 5,000 mapped transcripts.
Supplementary Table 6 | TB Cluster Enrichments. We examined sets of enriched sets of genes among all cells (irrespective of TB exposure) within Cluster 1, 2 and 3 using the find.markers function in Seurat, which implements a ‘roc’ test to identify relative expression differences. (a) From this analysis, we identified sets of genes exclusively enriched in each cluster but not the others. For each of the identified gene sets, we performed gene set enrichment analysis in DAVID and GSEA. For analysis in DAVID, we compared each gene list for enrichment among GO terms and curated pathways against a background list of 9045 genes contained in the DAVID database that were detected in at least 5% of cells. For analysis using GSEA, we compared each gene list to the complete database of gene sets contained within GSEA. (b, c) Within Cluster 1, we observe unique enrichment of 134 genes related to TNF-alpha signaling, inflammation, immune response and LPS response among 1099 cells. (d, e) In Cluster 2, we observe exclusive enrichment of 251 genes among 904 cells that distinguish monocytes from dendritic cells in culture and characterize TNF-alpha signaling. (f, g) Finally, in Cluster 3, we observe unique enrichment of 118 genes among 557 cells related to specifically to hypoxia, LPS stimulation, TNF-alpha signaling and apoptosis.
Supplementary Table 7 | Cluster Enrichments between Exposure Groups. Initially, we separately identified enriched genes among exposed and unexposed cells within Clusters 1, 2 and 3 using the find.markers function in Seurat. In total, we identified 18 genes with conserved enrichment among TB exposed cells within Clusters 1, 2 and 3. For each cluster, we identified the set of genes enriched among exposed cells and unexposed cells within each cluster. (a) We identified 28 enriched genes that were conserved among exposed cells across clusters and 31 conserved genes among unexposed, of which 18 were conserved between exposed and unexposed. We identified 38 genes uniquely enriched among exposed cells in Cluster 1 and 54 genes uniquely enriched among unexposed cells, of which 5 were conserved between exposed and unexposed cells within Cluster 1. We identified 134 genes unique to Cluster 2 among exposed cells and 200 genes unique to Cluster 2 among unexposed cells, of which 35 were conserved between exposed and unexposed cells within Cluster 2. In Cluster 3, we identified 43 genes unique to cluster 3 among exposed cells and 44 genes unique to Cluster 3 among unexposed cells, of which 9 were conserved between exposed and unexposed cells within Cluster 3. We performed gene set enrichment analyses for single gene list using DAVID and GSEA. In DAVID, we specified a background list of 9045 genes and examined enrichments within GO terms and curated pathways. For the analysis in GSEA, we made comparisons of gene lists to the complete database of gene sets within GSEA. (b, c) Among the 18 genes conserved across clusters in both exposed and unexposed cells, we observed strong enrichment for LPS response, TNF-alpha signaling, phagosome formation and macrophage activation. (d, e) Among the 5 genes unique to Cluster 1 conserved between exposed and unexposed cells, we observed enrichment of PI3K-Akt signaling and immune activation. (f, g) Among the 35 genes unique to Cluster 2 conserved between exposed and unexposed cells, we observed enrichment of genes related to monocyte culture and the coagulation cascade. (h, i) Among the 9 genes unique to Cluster 3 conserved between exposed and unexposed, we observed enrichment of genes up-regulated by HGF and apoptosis.
Supplementary Table 8 | Differentially Expressed Genes between TB Exposed and Unexposed Cells within Each Cluster. We performed a likelihood ratio test to identify genes differentially expressed between TB exposed and unexposed cells within each cluster. (a) Differential expression results between 673 TB-exposed and 426 unexposed cells in Cluster 1. (b) Differential expression results between 627 TB-exposed and 277 unexposed cells in Cluster 2. (c) Differential expression results between 386 TB-exposed and 171 unexposed cells in Cluster 3.
Supplementary Table 9 | TB Infection by Cluster Enrichments. Initially, we performed a LRT within each cluster to identify genes differentially expressed between TB exposed and unexposed cells. For each cluster, we created lists of genes differentially expressed with p-values less than 5.0×10−6 within each cluster (Figure 3c). (a) We then compared these lists to identify genes that are differentially expressed genes exclusively within each cluster. We also identified a list of 37 genes detected as differentially expressed across all clusters. We then performed gene set enrichment analysis in DAVID and GSEA to examine functional enrichment of the identified gene sets (i.e. Genes conserved across and unique to each cluster). We performed analysis in DAVID, comparing genes 37 conserved genes, 22 genes unique to Cluster 1, 142 genes unique to Cluster 2, and 40 genes unique to Cluster 3 to a background list of 9,381 genes expressed in at least 5% of filtered cells (Methods). Using GSEA, we made comparisons of the above gene list to the complete list of curated gene sets within the GSEA database (MSigDB v5.1: http://software.broadinstitute.org/gsea/msigdb/index.jsp). (b,c) Within Cluster 1, we observed unique enrichment of genes related to growth, proliferation and cell cycle. (d, e) Within Cluster 2, we observed enrichment of genes that identify monocyte and dendritic cell culture in addition to proliferation. (f, g) Within Cluster 3, we observed unique enrichment of genes related to hypoxia, oxidative stress and oxygen homeostasis.
Supplementary Table 10 | GSEA Comparisons of Exposed and Unexposed Cells within Each Cluster. We performed comparisons between M. tuberculosis exposed and unexposed cells within each cluster using GSEA. For each cluster we created .gct files containing normalized expression data for every cells within each cluster and assigned phenotypes (i.e. TB exposed vs. unexposed) to each cell using a .cls file. We then performed gene set enrichment analysis for each cluster across the complete gene set database in GSEA with 1000 permutations of assigned phenotype. (a,b) In cluster 1, we observed enrichment of dendritic cell maturation, monocytes in culture, response to L. donovani, and TNF-alpha signaling among 673 TB exposed cells and relative enrichment of ribosomal genes and protein synthesis among 426 unexposed cells. (c,d) In Cluster 2, we observed enrichment of LPS response, dendritic cell maturation, IL1 stimulation and response to TGF-beta among 627 TB exposed cells and relative enrichment of housekeeping functions, ribosomal genes and translation among 277 TB unexposed cells. (e,f) In Cluster 3, we observed enrichment of among delayed response to LPS (48 response), TLR7/8 stimulation, inflammatory response, intracellular infection and TNF signaling among 386 TB exposed cells and relative enrichment of housekeeping functions (ribosome, translation, actin) among 171 unexposed cells. (g,h) In Cluster 4, we observed enrichment of mitochondrial gene signatures, oxidative phosphorylation, hypoxia response and interferon response among 74 TB exposed cells and enrichment of ribosomal genes and translation among 975 unexposed cells. (i,j) In Cluster 5, we observed enrichment of LPS stimulation, TNF signaling, sepsis and dendritic cells maturation among 988 TB exposed cells and enrichment of ribosomal proteins and translation among 41 unexposed cells.