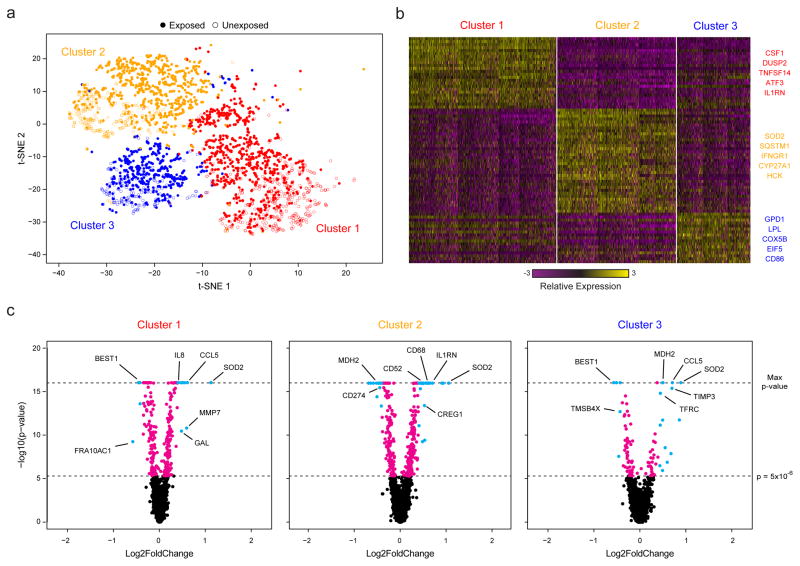
Figure 3. Sequencing of TB-Exposed Macrophages in a BSL3 Facility Using Seq-Well.
(a) t-SNE visualization of single-cell clusters identified among 2,560 macrophages (1,686 exposed, solid circles; 874 unexposed, open circles) generated using 5 principal components across 377 variable genes (Methods). (b) Marker genes for the 3 phenotypic clusters of macrophages highlighted in (a). (c) Volcano plots of differential expression between exposed and unexposed macrophages within each cluster showing genes enriched in cells exposed to M. tuberculosis. In each plot, a p-value threshold of 5.0 ×10−16 based on a likelihood ratio test was used to establish statistical significance, while a log2-fold change threshold of 0.4 was used to determine differential expression. Genes with p-values less than 5.0×10−6 are shown in cyan and absolute log2-fold changes greater than 0.4; In magenta are genes with p-values less than 5.0×10−6 but absolute log2-fold changes less than 0.4; and, in black, are genes with p-values greater than 5.0×10−6 and absolute log2-fold changes less than 0.4.