Abstract
Within nine dentin dysplasia (type II) and dentinogenesis imperfecta (type II and III) patient/families, seven have one of four net −1 deletions within the ~2kb coding repeat domain of the DSPP gene while the remaining two patients had splice-site mutations. All frameshift mutations are predicted to change the highly soluble DSPP protein into proteins with long hydrophobic amino acid repeats that could interfere with processing of normal DSPP and/or other secreted matrix proteins. We propose that all previously reported missense, nonsense, and splice-site DSPP mutations (all associated with exons 2 and 3) result in dominant phenotypes due to disruption of signal peptide-processing and/or related biochemical events that also result in interference with protein processing. This would bring the currently known dominant forms of the human disease phenotype in agreement with the normal phenotype of the heterozygous null Dspp (−/+) mice. A study of 188 normal human chromosomes revealed a hypervariable DSPP repeat domain with extraordinary rates of change including 20 slip-replication indel events and 37 predominantly C-to-T transition SNPs. The most frequent transition in the primordial 9-bp DNA repeat was a sense-strand CpG site while a CpNpG (CAG) transition was the second most frequent SNP. Bisulfite-sequencing of genomic DNA showed that DSPP repeat can be methylated at both motifs. This suggests that, like plants and some animals, human methylate some CpNpG sequences. Analysis of 37 haplotypes of the highly variable DSPP gene from geographically diverse people suggests it may be a useful autosomal marker in human migration studies.
Keywords: dentin sialophosphoprotein, dentinogenesis imperfecta, dentin dysplasia, CpNpG methylation, epigenetics, slip-replication
INTRODUCTION
Autosomal dominant genetic diseases of dentin are historically classified by clinical and radiographic information into two categories: dentinogenesis imperfecta (DGI types I, II, and III; MIM] 125490 and MIM] 125500) and dentin dysplasia (DD types I and II; MIM] 125400 and MIM] 125420) [Shields et al., 1973]. Patients with DGI typically have amber-brown, opalescent teeth that fracture and shed their enamel during mastication, thereby exposing the dentin to rapid wear. Radiographically, the crown appears bulbous and pulpal obliteration is common. While the primary teeth in DD II appear phenotypically identical to DGI, the permanent teeth typically show normal to mild discoloration and will often have thistle-shaped pulps and pulp stones. The overlapping clinical findings between these two diseases suggest a continuum of phenotypic findings, consistent with linkage of genetic loci for both DGI II and III and DD II to a common interval of chromosome 4q21 [Ball et al., 1982; Boughman et al., 1986; Dean et al., 1997] and specifically to the dentin sialophosphoprotein (DSPP) gene (NM_014208) [Hart and Hart, 2007].
Genetic studies of DSPP have been hampered by the difficulty in sequencing the highly repetitive region of DSPP exon 5 [Kim and Simmer, 2007]. A phenotype similar to DGI was reported in the DSPP (−/−) homozygotic knockout mouse [Sreenath et al., 2003]. Curiously, unlike the dominant phenotype reported in the human heterozygotic null DSPP (1/−) [Song et al., 2006; Zhang et al., 2001], there were no obvious tooth abnormalities found in the heterozygous (1/−) DSPP knockout mice (A.B. Kulkarni, Functional Genomics Unit and Gene Targeting Facility, NIDCR, NIH, Bethesda, MD; personal communication). The DSPP gene, originally thought to be expressed solely by odontoblasts in the formation of dentin [MacDougall et al., 1997a], was later found to be expressed at much lower levels in bone [Qin et al., 2002] and many metabolically active ductal epithelial cells [Ogbureke and Fisher, 2004, 2005, 2007]. DSPP consists of five exons, the first of which is noncoding. Exons 2–5 encode the 1,300–amino acid DSPP protein, which is generally accepted to be cleaved into two fragments, dentin sialoprotein (DSP) and dentin phosphoprotein or phosphophorin (DPP) [George et al., 1999; MacDougall et al., 1997b]. DSPP is the largest member of the SIBLING family of genes, each of which contains the integrin-binding tripeptide, RGD [Fisher and Fedarko, 2003]. DSPP is unique among the SIBLING members in that exon 5 contains over 200 tandem copies of a nominal 9-basepair (bp) repeat encoding a series of tandem Ser Ser Asp repeats. In dentin, these 400 serines are thought to be phosphorylated, making DSPP one of the most negatively charged, hydrophilic proteins known in humans. The repeat portion of DSPP is relatively conserved at the amino acid level among all mammals although its length is quite different among species. However, the underlying DNA sequences encoding the DSPP repeat has been anecdotally reported to be variable among humans [Kim and Simmer, 2007]. Some of this variation may arise from the CpG and CpNpG motifs within the 9-bp DNA repeat unit. Methylated cytosines in these motifs can deaminate, resulting in stable C-to-T (or G-to A in the complementary strand) transitions at rates generally accepted to be about 10 times that of other base changes [Pfeifer, 2006].
In this study, we present the first comprehensive analysis of all five DSPP exons in individuals with DD II and DGI II and III. Several novel mutations in the DSPP gene that correlate with both DGI and DD phenotypes are identified and a hypothesis unifying the apparent discrepancies between the human (dominant) and mouse (recessive) forms of the diseases is proposed. Finally, the repeat portion of DSPP is characterized in 94 humans from 10 geographically distinct locations to define the hypervariable repeat region that reveals its potential utility in CpG and CpNpG methylation/deamination events as well as in human migration studies.
MATERIALS AND METHODS
Kindreds and Genomic DNA
A total of nine probands/kindred with nonsyndromic, autosomal dominantly-inherited dentin defects were available for this study. Two families segregated DD II and seven families segregated DGI II/III. Members of the Brandywine DGI cohort have been reportedly linked to chromosome 4q21 [Boughman et al., 1986] and an exon 2 mutation in a single member of this extended family (Family DGI-1) has been preliminarily reported [Hart and Hart, 2007] but the clinical information, DSPP repeat sequence, and haplotype data were not reported. One DD II family (Family DD-2) has also been clinically described and linked to chromosome 4q21 [Dean et al., 1997]. DNA was available for 22 members (15 affected and seven unaffected) from these nine kindred. Clinical photographs and radiographs on all patients were reviewed (by J.T.W. and T.C.H.). Affected individuals and family members were identified at dental clinics at the following locations: National Institutes of Health, Bethesda, MD; University of North Carolina, Chapel Hill, NC; University of Colorado School of Dental Medicine, Aurora, CO; and the University of Indiana School of Dentistry, Indianapolis, IN. Informed consent was obtained according to the Declaration of Helsinki and approved by the corresponding Institutional Review Boards. Genomic DNA was isolated from whole blood samples using standard protocols. Normal human genomic DNA was purchased from Coriell Cell Repositories (Camden, NJ): African American (HD04), Africans South of the Sahara (HD12), Amerindian Yucatan population (GM10970–GM10979), Ami tribe of Taiwan (HD25), Andes of South America (HD17), Atayal tribe of Taiwan (HD24), Caucasian (GM17208–GM17224), Chinese (HD32), Japanese (HD07), Mexican Indian (HD28), and Northern European (HD01). Genomic DNA was isolated from the HSF-6 human embryonic stem cell line (passage 69) (a gift from Dr. Pamela Gehron Robey and Dr. Sergei Kuznetsov of the NIDCR/NIH) using standard protocols. The mutation nomenclature conforms with journal guidelines (www.hgvs.org/mutnomen). All numbering assumes the A of the ATG start codon (codon 1) as nucleotide 1. Reference sequence: GenBank NM_014208.3.
DSPP Mutation Analysis in Early Exons
The first four exons and the nonrepeat 50 portion of exon 5 of DSPP were amplified using the primers listed in Supplementary Table S1 (available online at http://www.interscience.wiley.com/jpages/1059-7794/suppmat). Briefly, 100 ng of DNA was amplified using standard PCR conditions with an annealing temperature of 581C for 40 sec. Following PCR, products were either gel-purified using the GeneClean kit (Qbiogene, Santa Ana, CA) or using exonuclease (Epicentre, Madison, WI) and shrimp alkaline phosphatase (USB, Cleveland, OH) according to the manufacturers’ instructions. The purified amplicons were sequenced in both directions using Big Dye Terminator v3.1 (Applied Biosystems, Foster City, CA) chemistry and a 3730 DNA analyzer (Applied Biosystems) by the NIDCR Division of Intramural Research DNA Sequencing Core. Alignments were constructed using the basic alignment search tool (BLAST; www.ncbi.nlm.nih.gov/blast) and the DSPP reference sequence NM_014208.
Haplotype Analysis for Families with c.2525delG Mutation
Five STR markers (D4S1534, D4S2409, D4S2929, D4S2284, and D4S2460) that flank the DSPP gene were genotyped using standard methodology [Hart et al., 2003]. Two additional sets of primers were used to genotype known intragenic SNPs, using the profile listed for DSPP mutation analysis, but with an annealing temperature of 601C for 30 sec. Primer set 7 (Supplementary Table S1) includes rs2736978, rs34603924, and rs2615487. Primer set 9 (Supplementary Table S1) includes rs2627699, rs2736980, and rs2846914. PCR products were electrophoresed through 1.8% agarose gels, bands (497 bp for set 7; 564 bp for set 9) were extracted using the GeneClean kit, and sequenced as described above.
Cloning and Sequencing of DSPP Repeat Domain
Because of the difficulty in sequencing the DSPP repeat within exon 5, the full-length 2.4-kb repeat domain (DPP) as well as the 1.2-kb 30 “hypervariable repeat region” (DSPP HVRR) were each cloned. Plasmid DNA from at least 10 colonies for each cloning event were necessary to verify a nominal 1:1 ratio of the two alleles and to distinguish true SNPs from occasional Taq DNA polymerase-generated changes. For the PCR of the HVRR, the forward primer is based on a unique Arg codon at basepair number 2692 located in the middle of the Ser Ser Asp repeat domain. In over 250 chromosomes analyzed, only one haplotype (DSPP HVRR Hap 37 GenBank accession no. EU278676) lacked this Arg and was analyzed using unusually long sequencing reactions of DPP alone.
Genomic DNA (300 ng) was used to amplify DPP and DSPP HVRR with 1 Platinums Taq PCR buffer, two units of Platinums Taq DNA polymerase (Invitrogen), 0.1mM of each dNTP, 1.5mM MgCl2, and 0.2 mM each of forward and reverse primer (Supplementary Table S1) in a total volume of 100 ml. The PCR amplification protocol for DPP was as follows: 941C for 5 min; 35 cycles of 941C for 30 sec, 551C for 30 sec, and 3 min at 721C; and then a final 5 min at 721C. The PCR amplification protocol for DSPP HVRR was as follows: 941C for 5 min; then 40 cycles of 941C for 30 sec, 651C for 30 sec, and 1.2 min of 721C; and then a final 5 min at 721C. PCR products (2.0–2.4 kb for DPP and 1.0–1.2 kb for DSPP HVRR) were excised from 1% agarose/TBE gels, purified using the Qiagen (Valencia, CA) Gel Extraction kit, and eluted in 30 ml of the provided elution buffer (EB). A total of 2 ml of gel-purified PCR cDNA was reacted with 0.5 ml Topos TA cloning vector (pCR4-TOPO; Invitrogen) in the presence of 0.5 ml of the provided salt solution, and incubated at room temperature for 30 min. A total of 20 ml of MAX Efficiencys Stbl2TM competent cells (Invitrogen) were transformed with 1 ml of the above reaction for 30 min on ice followed by 25 sec at 421C treatment, 2 min on ice, and incubation for 90 min with gentle agitation at 301C with 180ml of the provided SOC nutritional growth medium. Cells were spread on a 60-mm Luria broth (LB)- agar/ampicillin (100 mg/ml) plate and grown at room temperature for 72 hr (until colonies were greater than 1mm in size). Selected colonies were grown overnight in 5ml LB/ampicillin (100 mg/ml) at 371C with vigorous shaking. Plasmids were isolated from the entire culture using Promega’s (Madison, WI) Wizard SV Miniprep kits and eluted with 50 ml of water. DNA sequencing was performed using M13 forward and reverse primers. DPP was also sequenced with an internal forward oligonucleotide (CAGACAGCAGCAAATCAGAG). Most sequencing reactions were done with 30 cycles; however, low amounts of plasmid required more cycles.
Bisulfite Treatment for Identification of 5-Methyl-Cytosines in DSPP Repeat
A nonmethylated control DPP PCR product (Coriell African American genomic DNA, GM17031) was added to aliquots of Ssp-I digested genomic DNA (see below) from human embryonic stem (ES) cells and a somatic cell source (Coriell Northern European, GM17007); both sets were treated with bisulfite to determine the presence of 5-methyl-cytosines. Due to a homozygotic indel in the nonmethylated PCR-product control DNA, which was absent in the experimental genomic DNA samples, the sequencing results could be distinguished as control or genomic DNA. Standard bisulfite conditions described in the literature were insufficient to completely convert all of the PCR-product control DNA cytosines to thymidines. The final bisulfite method used was a variation on previously described methods [Paulin et al., 1998; Warnecke et al., 2002]. The repeat domain of DSPP within exon 5 is flanked by multiple Ssp-I restriction sites within the adjacent introns. Like the naturally occurring deamination events in vivo [Lindahl and Nyberg, 1974], bisulfite reactions are ineffective unless the DNA is single stranded during the process. Therefore, the experimental genomic DNA was digested with Ssp-I to render the target DSPP DNA smaller in size and therefore less likely to reform the double helical DNA after denaturation and also to be more similar in size to the control PCR template DNA. Approximately equimolar amounts of experimental genomic DNA and control PCR DNA were combined and treated under final conditions of: 1.72M sodium metabisulfite, 5.36M urea, and 0.5mM hydroquinone (all from Sigma, St. Louis, MO) in a thinwalled 200-ml PCR tube. The samples were heated to 941C for 2 min and then treated for 20 cycles of 941C for 30 sec and 551C for 60 min. The product was purified using the Qiagen PCR purification kit and eluted in 80 ml of EB. A total of 8 ml of 3M NaOH was added to 72 ml of the eluted DNA and incubated for 15 min at 371C, neutralized by 56 ml of 5M ammonium acetate, purified, and eluted with 30 ml EB. A NanoDrop spectrometer (Thermo Scientific, Waltham, MA) was used to quantify the final DNA concentration and 200 ng DNA was used as template for PCR. These samples were amplified as above using bisulfite forward and reverse primers (Supplementary Table S1) in a final volume of 50 ml. Cycle parameters: 5 min at 941C, 35 cycles of 30 sec at 941C, annealing for 30 sec at 501C, 20 sec 721C extension, with a final 5 min at 721C. Primer oligonucleotides were designed to amplify the last few repeats in the sense strand after the bisulfite reaction. The 200-bp amplicons were cloned into TOPO TA cloning vector and sequenced as described above.
Sequence Analysis
Construction of the DPP sequence for each allele was completed by compiling data from both the DSPP HVRR and DPP sequencing results. Sequences were assembled using SequencherTM 4.7 (Gene Codes Corporation, Ann Arbor, MI) software and the online version of the CAP3 Sequence Assembly Program (http://deepc2.psi.iastate.edu/aat/cap/cap.html) [Huang and Madan, 1999]. Final results were verified by visual analysis. The alignment of the 37 human DSPP HVRR haplotypes from the Coriell samples was completed by hand and entered into Networkr 4.2.0.1 (Fluxus Technology, Ltd., Suffolk, UK) to compose the phylogenetic network using the reduced median network calculation option. Hydrophobicity was determined by generating a Kyte-Doolittle plot (www.vivo.colostate.edu/molkit/hydropathy/index.html) [Kyte and Doolittle, 1982]. The plots were constructed with serines instead of phosphoserines; the latter would have resulted in even stronger hydrophilic scores for the nonmutated portions of the DSPP repeat domain.
RESULTS
Patient Description and Mutational Analysis
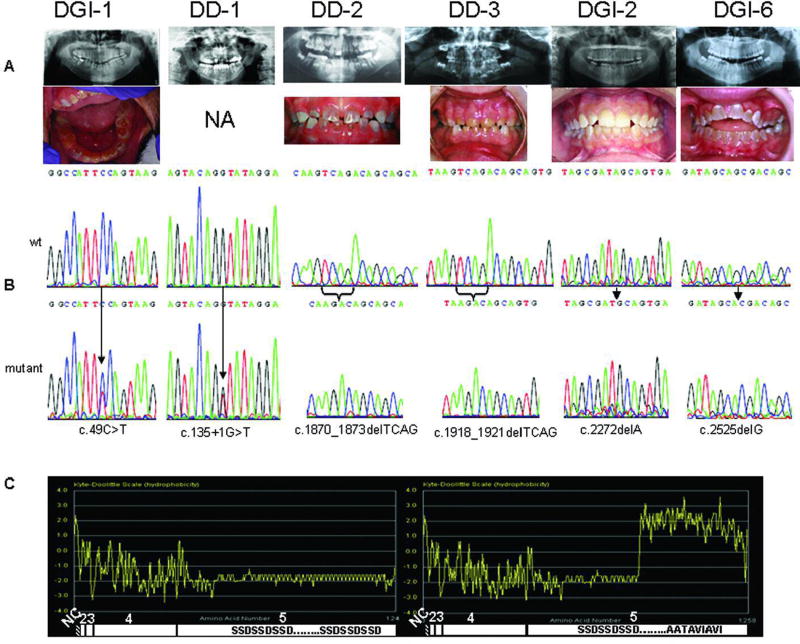
As shown in Figure 1A and Table 1, a cohort of nine patients/families diagnosed with either DGI or DD were sequenced for DSPP mutations. Two probands, Patients DGI-7 and DGI-1, had mutations identified in the nonrepeat portion of DSPP (Fig. 1B). Patient DGI-7 was found to be heterozygous for a c.13511G4T alteration that affects the splice donor site of intron 3. Interestingly, a different mutation of this same nucleotide, c.13511G4A, has been reported in a family with DGI-II [Xiao et al., 2001]. Proband DGI-7 (of the long-studied Brandywine cohort) was found to be heterozygous for a c.49C4T transition that is predicted to cause a p.P17S substitution. The identical mutation was recently reported in a Chinese DGI-II family [Zhang et al., 2007] and a c.49C4A transition (p.P17 T) was previously reported in a different Chinese DGI-II family [Xiao et al., 2001]. One affected member of the same Brandywine cohort was reported to have had a 36-bp in-frame deletion followed by an 18-bp inframe insertion within the 30 end of the repeat domain of DSPP [Dong et al., 2005]. We confirmed this pair of indels in Proband DGI-1 (DSPP HVRR Hap 38, Genbank accession no. EU278677) but contend this is likely a normal variant and that the c.49C4T alteration is likely the causative event (see below).
Figure 1.
TABLE 1.
Clinical Description and Mutations Identified in this Study*
| Family | Clinical Description | Mutation |
|---|---|---|
| DGI-7 | Classic DGI | c.13511G4T |
| DD-2 | Primary teeth appear opalescent; histologically the primary teeth showed coronal mantle dentin that was more regular compared to circumpulpal dentin and the dentinal tubules in the circumpulpal dentin were sparse and highly branched; permanent teeth exhibited normal histology | c.1870_1873delTCAG |
| DD-3 | Classic DD | c.1918_1921delTCAG |
| DGI-1 | Teeth appear opalescent-brown with extensive loss of enamel from occlusal surfaces. Underlying exposed dentin is reddish-brown. Radiographically, anterior and posterior teeth show cervical constriction and bulbous appearing crowns. Pulp chambers and root canals are obliterated, particularly for anterior teeth, pulps of posterior teeth are reduced in size. Periapical radiolucencies are associated with four posterior teeth and one anterior tooth. Patient reports no pain associated with any teeth. | c.49C4T |
| DGI-2 | Teeth appear clinically to have normal shape and size with an opalescent-amber appearance. Radiographically, posterior teeth show a cervical constriction with bulbous crowns. Most pulp chambers are obliterated, and crescent shaped pulps are evident in several molars. | c.2272delA |
| DGI-3 | Classic DGI | c.2525delG |
| DGI-4 | Classic DGI | c.2525delG |
| DGI-5 | Classic DGI | c.2525delG |
| DGI-6 | Classic DGI | c.2525delG |
Reference sequence NM_014208.
The remaining seven new probands were found to have frameshift mutations in the 2-kb repeat portion of DSPP exon 5. All seven showed a net −1 frameshift (i.e., a loss of 1 or 4 bp resulting in the same, new, −1 open reading frame) in the first half of the repeat (Fig. 2). The locations of these four mutations are reported with respect to the current DSPP reference sequence (NM_014208.3), numbering from the start codon (A is nucleotide 1). Because of the high degree of sequence variability among the normal DSPP haplotypes (see below), the values are not the exact numerical location of the mutation within the specific patient’s own DNA. The exact sequence for each unique frameshift is available in GenBank (accession no. EU284750–EU284753) and an alignment is available in Supplementary Table S2). Two different frameshift events were found in the two DD families. One DD family (Family DD-2) had a −4-bp (loss of TCAG) frameshift (c.1870_1873delTCAG; GenBank accession no. EU284750) while the other DD family (Family DD-3) had a similar 4-bp deletion 50 bp 30 (c.1918_1921delTCAG; GenBank accession no. EU284751). The two frameshifts were verified to be separate events when aligned with each other and to a normal control haplotype, due to the presence of identifiable sequences between the two mutation sites (Fig. 2; Supplementary Table S2). Interestingly, the TCAG lost in both DD families are two of the three times this precise sequence is found in the entire DSPP repeat, and these four bases may represent currently unexplained mutational hotspots in humans.
Figure 2.
Among five DGI patients/families, two different −1-bp deletions were found 30 to the DD frameshifts (Fig. 2). In a single DGI patient (Patient DGI-2), the loss of an A was identified (c.2272delA; GenBank accession no. EU284752) while in the other four families (Families DGI-3 to DGI-6), a G was deleted (c.2525delG; GenBank accession no. EU284753). There were no apparent unique qualities to the DNA sequence surrounding the deleted G to explain a mutational hotspot. All of the c.2525delG patients had the identical DSPP repeat haplotype (DPP Hap 2A; GenBank accession no. EU278627). Furthermore, detailed analysis of an 4-Mb portion of chromosome 4 surrounding the DSPP gene for all of the patients carrying the c.2525delG mutation identified in an identical haplotype (Supplementary Table S3). (A control sample with the same DPP Hap 2A haplotype but lacking the c.2525delG mutation was analyzed for comparison.) These data suggest the mutation identified in these four families was inherited identical by descent and is likely an example of a founder-effect mutation that has been propagated within the U.S. population. For every case in which DNA from family members was available, one normal and one frameshift mutation allele was found in each affected family member, and unaffected relatives had two normal alleles. In addition, no frameshifts were identified within the full DSPP repeat domain in 100 chromosomes from normal individuals of diverse geographic locations. The 2-kb repeat portion of DSPP in humans is the result of 220 tandem repeats of the nominal sequence, AGC AGC GAC. Each repeat encodes a Ser Ser Asp (SSD) tripeptide, although the exact number of the tandem repeats and the specific sequence of each underlying repeat have drifted to some degree since an apparent ancient expansion event(s). All of the −1 and −4 frameshifts would change the long, hydrophilic (phosphorylated) SSD repeats into a polypeptide rich in the hydrophobic amino acids valine (Val), alanine (Ala), and isoleucine (Ile). The purely hydrophilic and soluble DSPP protein therefore abruptly changes into a protein with an essentially hydrophobic carboxyterminal domain starting at the frameshift location and continuing past the normal stop codon until an in-frame stop codon is reached 12 codons later (Fig. 1C). The most 50 frameshift mutation (c.1870_1873delTCAG) in Family DD-2 would have 4600 hydrophobic amino acids at the carboxy-terminus. The slightly more 30 mutation (c.1918_1921delTCAG) in Family DD-3 would have a similar number. The DGI patients in our cohort had frameshifts that would translate into 500 (c.2272delA) and 400 (c.2525delG) hydrophobic amino acids.
Identification of Normal DSPP Haplotypes and Their Possible Use in Geo-Ethnic Studies
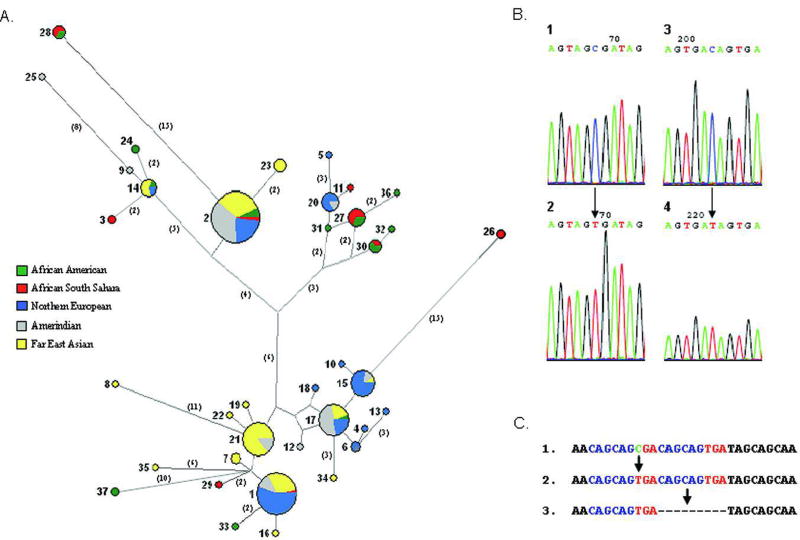
The entire DSPP repeat (DPP) was sequenced from 50 unaffected control individuals (100 chromosomes) of geographically diverse origins. Although, no frameshifts were observed in any control DSPP genes, an extraordinary amount of variation was identified within the 2.2-kb repeat domain, more similar to what is seen in microsatellite repeat polymorphisms. For example, there was a net difference of 432 bp (144 amino acids) between the longest (DSPP HVRR Hap 24; GenBank accession no. EU278663) and shortest (DSPP HVRR Hap 33; GenBank accession no. EU278672) haplotypes due to indels. It was also evident that within the entire repeat domain of DSPP, the majority of the differences occurred in the 30 half of the repeat. This 1.2- kb HVRR has somewhat arbitrarily been denoted to begin at a unique Arg (2,692 bp downstream from the start codon; GenBank accession no. NM_014208.3) and continues until only a few basepairs before the stop codon. Because the majority of differences in the DSPP gene in humans occurred in the HVRR portion, this 1.2-kb domain was analyzed further to define haplotypes within a population of control individuals. A comparison of 188 chromosomes from 94 individuals from 10 geographically distinct regions identified 37 unique haplotypes of DSPP HVRR (DSPP HVRR Hap 1–37; GenBank accession no. EU278640–EU278676, respectively). The 37 SNPs and 20 indels, which in specific combinations defined the individual haplotypes, are summarized in Supplementary Table S4A and B, respectively A complete alignment of all of the DSPP HVRR haplotypes is available (Supplementary Table S5).
Similar to the findings in human mitochondrial DNA and Y chromosome genes, common haplotypes of autosomal DSPP HVRR were present in all the studied ethnic groups from Coriell Cell Repositories [Maca-Meyer et al., 2001; Tishkoff and Verrelli, 2003; Underhill et al., 2001]. Other haplotypes appeared to be more limited to peoples sharing hypothesized human migration routes often associated with out-of-Africa theories (Fig. 3A). For example, haplotype 21 was very frequent in the Asian-descended groups (21% of the 100 chromosomes obtained from Chinese, Japanese, Taiwanese, and their migrant relatives, the Mexican and Andes groups) but was absent in the 88 chromosomes of African (African South Sahara and African American) and European (Caucasian and Northern European) descent (Fig. 3A). The cluster including haplotypes 27, 30, 31, 32, and 36 was only observed in our African populations. While analysis of the HVRR is the most simple and data-rich approach, additional information can be obtained by analyzing the entire DSPP repeat. Many of the most common, shared DSPP HVRR haplotypes could be further separated into one or more related but distinct haplotypes based on indels and SNPs that occurred 50 to the HVRR (DPP; GenBank accession no. EU278619–EU278639). For example, with the addition of the 50 SNPs/indels, two of the most frequent and otherwise shared HVRR haplotypes (1 and 17) separated along ethnic group lines: the Chinese and Japanese-associating alleles separating from the Caucasian, Northern European, and Amerindian groups for both of these haplotypes (GenBank accession no. EU278619–EU278624 for Hap 1 and EU278630–EU278631 for Hap 17).
Figure 3.
Origin of the DSPP Repeat Expansion and Evidence for Subsequent SNP Formation
Due to the repetitive nature of DSPP’s exon 5 and the large number of variants among humans (even more so when compared to our closest living relative, the chimpanzee; data not shown), the use of a standard outgroup was not a practical method to directly decipher the common ancestral sequences of the entire DSPP repeat. The current human population is generally agreed to have descended from a relatively small group of genetically narrow individuals that survived one or more severe population bottlenecks within the last few hundred thousand years or less [Liu et al., 2006]. Therefore, a significant number of the differences in the HVRR observed among the 94 individuals studied can reasonably be hypothesized to be predominantly the result of relatively recent changes in the DNA. When the SNPs of the HVRR were tabulated (Supplementary Table S4A), 75% of the changes could logically be concluded to be the result of C-to-T transitions (or G-to-A in the antisense strand). These nucleotide changes were usually at a CpG or a CpApG, suggesting that at least the repeat portion of the gene may be methylated and therefore susceptible to the relatively rapid deamination transition of 5-methyl-cytosine to thymine.
To measure the frequency of transitions within the entire DSPP HVRR and not just the SNPs observable between modern haplotypes, we quantified the changes in each consecutive repeat unit compared to that of a proposed primordial repeat sequence of AGC AGC GAC. This hypothesized primordial repeat is based on the following observations for a representative human DSPP sequence (NM_014208 from basepair 1765 to 3897): 1) Most of the serines in the repeat are either AGC or AGT codons, with 1% of the TCN type. There were only three TCN serine codons (TCA) in the entire human 2.2-kb DSPP repeat and these are all found in the most 50 portion of the sequence. TCN-type codons are not rare-usage codons for human proteins; indeed, they are not even rare in the nonrepeat coding portions of DSPP. This suggests that the majority of the DSPP repeat (and all of the HVRR) was likely due to a series of expansions involving a short repeat that lacked any TCN-type serine codons. 2) C-to-T transitions can be considered predominantly unidirectional due to deamination events (often due to 5-methyl-cytosine); therefore, T-to-C transitions are rare compared to C-to-Tevents. Thus AGTserines within the DSPP repeat are likely to have derived from AGC codons. A similar argument can be made for the silent change in the aspartic acid codon (GAC to GAT transitions). The GAC was probably the primordial codon and the GATs present are a result of later C-to-T transitions. 3) The nonomer repeats observed are GC-rich (i.e., AGC AGC GAC and not including AGT or GAT codons) and are more likely to be remnants of a primordial sequence expansion than to have been assembled from rare T-to-C type changes. Therefore, we propose that AGC AGC GAC is the primordial nonomer and that almost all of the current variations on this repeat in humans (particularly within the HVRR) are the result of C-to-T transitions (or G-to-A from the antisense strand). Using this primordial repeat as a simple outgroup, the relative changes in specific positions within the consecutive nonomers throughout the entire repeat can be estimated. Only complete nonomers within the repeat were used in these analyses. Because the processes of expansion and contraction of the repeats by slipreplication can occur using any set of two or more neighboring, usually identical, nonomers, the absolute number of changes in any one position over the entire repeat may not be particularly insightful. Finally, although the HapMap (www.hapmap.org) shows the DSPP repeat domain to be a recombination coldspot and we saw no direct evidence of recombination events (i.e., new haplotypes that logically are the recombination of common haplotypes), such processes cannot be ruled out as having occurred in the past. Considering all of the above, we determined the percent of change at each base position of the 9-bp primordial repeat compared to all other positions in the repeat unit (Table 2). Within DSPP HVRR, approximately two-thirds of second position AGC serine codons were found to be changed to the alternative serine codon, AGT. This high frequency of change fits well with CpG methylation and subsequent higher rates of deamination of the 5-methyl-cytosine in AGC(pG). Bisulfite sequencing analyses of somatic cell genomic DNA did confirm the presence of methylated cytosines in the repeat’s second serine CpG site just 50 to the stop codon (Fig. 3B-1). Bisulfite sequencing of nonmethylated (PCR) control DSPP DNA added to the same reaction (but recognizable as control due to the presence of an indel in the original template DNA) demonstrated that the bisulfite treatment conditions were sufficiently stringent to result in 100% loss of cytosines in all control reactions (Fig. 3B-2). (Paradoxically, the methylation of a cytosine that makes it more likely to change into a thymidine in normal biology, protects it from deamination into a thymidine in a bisulfite reaction.) The complementary CpG in the antisense strand results in a GAC-AAC (Asp to Asn) codon change in the sense strand. This change occurred in 16% of the repeat units found in DSPP HVRR, significantly lower than the CpG on the sense strand (Table 2). Asparagine amino acids appear to be tolerated periodically along the coding repeat, but such substitutions may be limited in spacing or in total number in order for the DSPP protein to function properly. Methylation-mediated deamination of CpG dinucleotides to TpG in vivo is significantly affected by the identity of the adjacent 30 base. Krawczak et al. [1998] have shown that CGA-to-TGA transitions in the noncoding strand (as in the second position Ser-to-Ser transition described above) occur about twice as frequently as the CGC-to-TGC transitions (as in the antisense transition resulting in Asp-to-Asn changes) in humans. Therefore, the lower percent of the antisense CpG transition (16% vs. 64%) may be due to a combination of lower rates of CpGpC methylation/deamination events and a moderate selection pressure against the occurrence of too many Asn amino acids within the DSPP repeat.
Table 2.
Percent of nucleotide change at each base position of the 9bp primordial repeat compared to all other positions in the repeat unit
| Primordial repeat unit: | A | G | C | A | G | C | G | A | C |
|---|---|---|---|---|---|---|---|---|---|
| Percent change in DPP (%) | 0 | 1 | 10 | 1 | 6 | 66 | 19 | 1 | 40 |
| Percent change in HVRR (%) | 0 | 1 | 6 | 1 | 2 | 64 | 16 | 1 | 35 |
The second most frequent change in the primordial repeat was a GAC(AG)-GAT(AG) transition. More than one-third of GAC codons had undergone a translationally silent change to GAT (Asp-to-Asp) throughout DSPP HVRR (Table 2). This is a much higher percent of change than seen in the other repeat’s non-CpG cytosines. This SNP could be explained by CpNpG methylation/ deamination events known to be common in plants and more recently reported in mouse ES cells [Dodge et al., 2002]. Sequencing data from bisulfite-treated human ES cell and somatic cell genomic DNA showed the occasional presence of CpNpG methyl-cytosines in this GAC codon near the end of the repeat (Fig. 3B-3). No GAC methyl-cytosines were detected in the PCR control samples (Fig. 3B-4). Other CpNpG sequences exist in the nonomer repeat unit, but these were not observed to change in the repeats as frequently nor were cytosines observed at these positions after bisulfite treatments in either ES cell or somatic cell genomic sense-strand DNA, suggesting that these sites are rarely, if ever, methylated. This low number of changes supports the previous observation that CpT methylation occurred much less often than CpA methylation in the mouse ES cells [Meissner et al., 2005]. Finally, the three adenosines within the repeat were the least likely to change (0–1%, Table 2).
Indel Formation Within the DSPP Repeat Domain
The nominal 9-bp DSPP repeat unit was prone to the formation of indels, most likely due to slip-replication events. Indels identified in the normal samples within the HVRR occurred in multiples of nine nucleotides (9, 18, 27, …) up to 189 nucleotides in the largest apparently single indel event. (Some indels within DPP [50 to the HVRR] are multiples of 3 bp and not the typical 9 bp seen in the HVRR.) The indels always resulted in an in-frame final product in the normal population, although the precise location of any one event (to within a single base) and/or the exact sequence of the bases physically inserted or deleted could not always be unambiguously assigned. In all cases, however, a narrow region for the slip-replication event was clearly indicated by the simplest alignment. In our analysis, the number and location of the indels were set to minimize the total number of possible haplotypes. Evidence of the requirement to often have two identical, neighboring repeats for a slip-replication event to occur is demonstrated when comparing all human DSPP HVRR haplotypes at nucleotide position 2861–2877 (Fig. 3C). Each of the identified haplotypes had one of the three sequences. In Figure 3C-1, the two nonomers are not identical; however, upon SNP formation (Fig. 3C-2) they became identical and perfectly situated for an in-frame slip replication (Fig. 3C-3). Most of the 20 indels identified in the 37 HVRR haplotypes had a clear pattern of slip-replication similar to this example.
DISCUSSION
As summarized in Table 3, reports over the past few years have disclosed several missense mutations in the first two coding exons of the DSPP gene, as well as several apparent splice junction mutations involving exon 3, which are all strong candidates for causing at least some cases of DD and/or DGI. A single nonsense mutation at the 30 end of exon 3 has also been described in two different families [Song et al., 2006; Zhang et al., 2001]. We think it is important to note that no nonsense mutations among the dominant human dentin diseases are currently known within the last two exons of DSPP, which together comprise 96% of the coding sequence. About half of the DSPP-coding domain has not been studied in the majority of patients because it is composed of over 200 tandem copies of a nominal 9-bp repeat. Analysis of this 2-kb repeat requires cloning of individual alleles because of the large number of sequence polymorphisms among the numerous haplotypes. The many indels interfering with the interpretation of direct PCR sequencing of both alleles simultaneously and the paucity of unique internal sequences that can successfully be used as primer sites are both obstacles that have kept researchers from successfully finding mutations in this region. We present the first comprehensive analysis of the DSPP gene, including the repeat domain in a cohort of seven DGI and two DD patients/families. With the exception of the c.49C4Tmutation preliminarily reported for Family DGI-1, the remaining eight probands were heterozygous for 1 out of 5 novel mutations, including four different frameshifts in the highly repetitive portion of exon 5. Results of our study cohort suggest that a significant portion (and perhaps the majority) of DSPP mutations associated with nonsyndromic DD and DGI are likely to be found in the repetitive portion of exon 5. Interestingly, there is currently a trend in this data that DD patients have more 50 frameshift mutations (and therefore more hydrophobic amino acids) than the DGI patients, but a larger number of cases need to be studied to determine if these observations are causative for the two currently distinguishing phenotypes or if DD and DGI represent an overlapping spectrum of DSPP mutations.
TABLE 3.
Mutations in DSPP Resulting in Dentin Defects
| cDNAa | Proteinb | Mutation class | Ethnicity | Diagnosis | References | |
|---|---|---|---|---|---|---|
| Exon 2 | c.16T4G | p.Y6D | Signal peptide | Caucasian (German)c | DD-II | Rajpar et al. [2002] |
| c.44C4T | p.A15V | Signal peptide | Central American | DGI-II | Malmgren et al. [2004]] | |
| c.49C4A | p.P17T | IPV mutation | Chinese | DGI-II | Xiao et al. [2001] | |
| c.49C4T | p.P17S | IPV mutation | Brandywine triracial isolate | DGI-II | Hart and Hart [2007] | |
| Chinese | DGI-II | Zhang et al. [2001] | ||||
| Intron 2 | c.52-3C4G | p.V18_Q45del | IPV mutation | Korean | DGI-II | Kim et al. [2004] |
| c.52^3C4A | Finnish | Holappa et al. [2006] | ||||
| Exon 3 | c.52G4T | p.V18_Q45del or p.V18F | IPV mutation | Chinese | DGI-II | Xiao et al. [2001] |
| Korean/Caucasian | DGI-III | Kim et al. [2005] | ||||
| Chinese | DGI-III | Song et al. [2006] | ||||
| Finnish | DGI-II | Holappa et al. [2006] | ||||
| c.133C4T | p.V18_Q45del or p.Q45X | IPV mutation | Chinese | DGI-II | Zhang et al. [2001] | |
| Chinese | Song et al. [2006] | |||||
| Intron 3 | c.13511G4A | p.V18_Q45del | IPV mutation | Chinese | DGI-II | Xiao et al. [2001] |
| c.13511G4T | Caucasian | DGI-II | This study | |||
| Exon 5 | c.1870_1873delTCAG | p.S624TfsX687 | Frameshift | Caucasian | DD-II | This study |
| c.1918_1921delTCAG | p.S640TfsX671 | Frameshift | Caucasian | DD-II | This study | |
| c.2272delA | p.S758AfsX554 | Frameshift | Caucasian (Northern European) | DGI-II | This study | |
| c.2525delG | p.S842TfsX471 | Frameshift | Caucasian (Northern European) | DGI-II | This study |
All numbering assumes the A of the ATG start codon as nucleotide number one. Reference sequence NM_014208.3.
These are predictions only and not verified consequences.
Personal communication with Dr. M.J. Dixon
All of the frameshift mutations (loss of 1 or 4 bp) caused the reading frame to change from tandem hydrophilic (and phosphorylated) Ser Ser Asp repeats to long stretches of hydrophobic amino acids rich in Val, Ala, and Ile. What was once perhaps the most hydrophilic and acidic protein found in humans becomes essentially hydrophobic in character for several hundred amino acids 30 to the frameshift mutation. At this time it is not known whether the mutant proteins are fully processed and secreted out of the odontoblast or if they form insoluble aggregates in the rough endoplasmic reticulum (rER) or subsequent processing organelles. If not properly processed, the mutant DSPP may accumulate within the synthesis or secretory apparati of the cell and cause a reduction in the production of the DSPP derived from the wildtype allele. This may cause the amount of DSPP in the dentin to be substantially lower than predicted by haploinsufficiency and may explain the dominant effect of these mutations.
DSPP and/or its two major breakdown products, DSP and DPP, are the most abundant protein (after type I collagen) in dentin and large amounts of insoluble mutant DSPP within the protein synthesis/modification pathways of the odontoblasts could reasonably be hypothesized to interfere with the processing of collagen or other critical proteins and thereby result in the dominant phenotypes of DGI and DD. This model can logically link these net −1-bp frameshift mutations to another major class of autosomal dominant dentin disorders, osteogenesis imperfectaassociated DGI (OI-DI or DGI I). (Interestingly, DGI is not found in cases of simple haploinsufficiency of a collagen allele such as type-I OI; Hart and Hart [2007]). In cases of OI due to dominant negative mutations, DGI is frequently present and may be the most penetrant finding [Pallos et al., 2001]. In DGI I, either collagen IA1 or IA2 is mutated, resulting in the formation and accumulation of incorrectly assembled collagen trimers within the cellular processing machinery [Delahunty and Bonifacino, 1995; Gajko-Galicka, 2002]. The matrix made under rapid growth conditions of childhood is poor in quality resulting in weak, opalescent teeth and easily broken bones. After puberty, many of these OI patients show a markedly lower rate of bone fracture, perhaps because the rate of bone formation slows sufficiently to allow the cell to handle the incorrectly assembled collagen trimers and permit the orderly accumulation of a matrix more closely resembling that of normal tissue [Marini, 1988; Byers and Cole, 2002]. Under conditions of rapid matrix synthesis, DSPP may precipitate or interact with the hydrophobic rER or Golgi membrane components, disrupting the synthesis and/or secretion of the normal collagen trimers and therefore preventing the assembly of adequate dentin matrices in rapidly growing teeth. The lack of any reported phenotype in metabolically active ductal epithelial tissues (e.g., kidney) and skeletal bone of heterozygotic DSPP mutant patients may be due to the much lower levels of DSPP expressed in these tissues and therefore proportionally lower levels of cell process-interfering mutant protein.
We propose that the two nonframeshift mutations found in this study, as well as all of the previously published mutations found in the DSPP gene (Table 3) (including the reported nonsense mutation c.133C4T), can be hypothesized to cause dominant forms of DD and DGI due to errors in signal peptide (or subsequent) processing events that result in scenarios similar to that described above for the −1-bp frameshift mutations. Like most secreted proteins, DSPP has a short, hydrophobic signal peptide that upon synthesis directs the ribosome/mRNA complex to a docking site on the rER. As translation of the remainder of DSPP continues into the lumen, the signal peptide is cleaved by one of a family of membrane-associated signal peptide peptidases (SPP). Two previously described missense mutations directly affect either the critical hydrophobicity of the signal peptide (p.Y6D; Rajpar et al. [2002]) or the requirement for a small amino acid on the amino-terminal side of the SPP site (p.A15V; Malmgren et al. [2004]). As discussed in previous literature, these changes in the properties of the signal peptides could result in incorrect processing of the mutated DSPP protein. Either the accumulation of the mutant protein in the cytosol due to the loss of the docking signal or the continued occupation on the ribosome/docking sites may indirectly result in insufficient processing of the normal DSPP allele or of other critical proteins such as collagen.
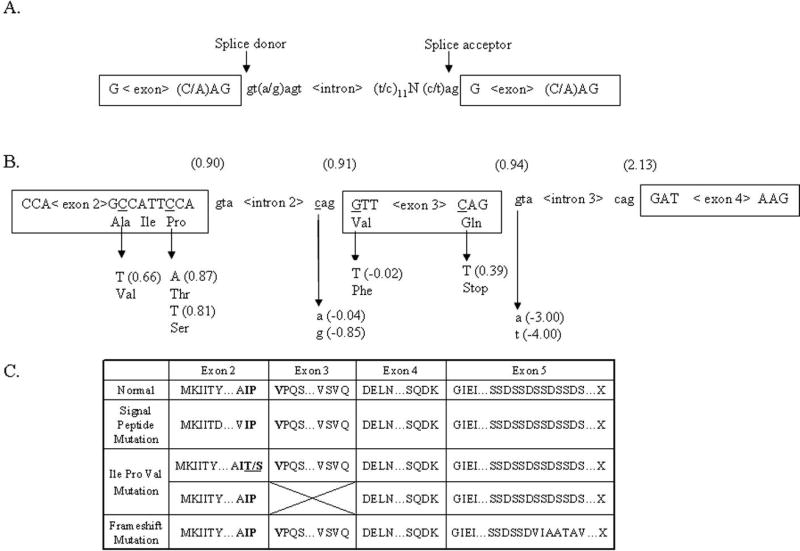
As a corollary, we further propose that the conservation of the chemical properties of the first three amino acids on the carboxyterminal side of the SPP cleavage site may also be critical for the efficient processing of the DSPP’s signal peptide (as is the case for many other proteases in biology). The first two amino acids of the mature protein (immediately after the SPP cleavage site) are normally encoded by the last six bases of exon 2 and the third by the first three bases of exon 3. These three amino acids in DSPP are Ile Pro Val (IPV), with the unique and structurally confined proline being flanked by two hydrophobic amino acids. With the exception of a chemically similar Val substituting for the Ile in the elephant, this tripeptide sequence is, to our knowledge, invariant within all animal species sequenced to date (University of California, Santa Clara (UCSC) Genome Bioinformatics; http://genome.ucsc.edu). The chemically similar motif is also found at the SPP cleavage site in several of the other major human proteins secreted during tooth matrix assembly, including: DMP1 (LeuProVal), OPN (LeuProVal), ameloblastin (ValProPhe), and amelogenin (MetProLeu). This completely conserved Pro in DSPP was shown to be mutated to a serine (p.P17S) in our single representative of the Brandywine DGI family (Family DGI-1), a result identical to that recently reported for a different kindred by Zhang et al. [2007], and similar to the p.P17 T first noted by Xiao et al. [2001]. These changes may cause signal peptide processing errors by the odontoblast SPP and result in dominant dentin diseases similar to that seen in direct changes in the signal peptide. At first, the change of the mature protein’s third position, Val, in the p.V18F (c.52G4T) event [Holappa et al., 2006; Kim et al., 2005; Song et al., 2006; Xiao et al., 2001] looks to be just a chemically conserved missense mutation unlikely to cause significant changes in the processing of DSPP. However, we propose that this represents the first in a series of splice-site mutations involving exon 3. The consensus sequence derived for the underlying DNA of the mRNA’s splice donors and acceptors is shown in Figure 4A. The GT and AG ends of the intronic sequence are nearly invariant, although GT and AG are common dinucleotide sequences. Therefore, the likelihood of a splice event occurring at any particular GT (GU in the RNA transcript itself) or AG increases as the sequences better fit the consensus. Using the SplicePort program (http://spliceport.cs.umd.edu), the p.V18F mutation changes the functional (10.91) splice acceptor site: CAG^GTT(Val) into a sequence predicted to be ineffective, CAG^TTT(Phe) (−0.02; see Fig. 4B) [Mount, 1982]. Due to the lack of a strong alternative splice acceptor site within 50 bp of the damaged one [Krawczak et al., 2007], the loss of this spice junction would likely cause exon 3 to be skipped, bringing exon 4 (with its normal splice acceptor, CAG^GAT) in-frame to exon 2. This would make the carboxy-terminal side of the SPP cleavage site become Ile Pro Asp, thereby replacing the highly conserved hydrophobic Val with a very hydrophilic amino acid, aspartic acid. Our splice-site mutation in Patient DGI-7, as well as all of the previously published splice junction mutations (Table 3), will result in this same loss of exon 3 and consequent replacement of the hydrophobic Val with a hydrophilic Asp. Even the published nonsense mutation pQ45X [Song et al., 2006; Zhang et al., 2007] can logically be hypothesized to frequently result in the loss of exon 3. The normal splice donor site for exon 3 (including the CAG codon for glutamine, which is conserved in all known species) is CAG^GT, but the transition event makes it TAG^GT, thereby changing a comparatively weak donor site (10.94) into a predicted less functional site (10.39; see Fig. 4B) that may cause exon 3 to be skipped at least some of the time. Using neural network–based splice-site recognition programs in the study of human disease-causing splice junction mutations, Krawczak et al. [2007] noted that many disease-causing mutations were the result of changes in donor splice-sites that were already suboptimal. Thus, all of the early exon missense, nonsense, and splice junction mutations can be hypothesized to be dominant due to errors of signal peptide (or subsequent IPV-dependent) processing events that may, in turn, result in the normal DSPP protein, collagen, or other critical proteins not being properly processed for the rapidly accumulating dentin matrix.
Figure 4.
Given that DSPP haploinsufficiency (1/−) in mice does not result in any dentin phenotype, our hypothesis naturally leads to a corollary which predicts that mutations in DSPPexons 2, 3, and 4 that do result in a true, single null allele in humans will be found in human recessive dentin disorders. It may also be possible that exon 5 nonsense mutations (including both point mutations and net 11 frameshift mutations in the repeat domain, which always quickly result in stop codons) as well as net −1 frameshift mutations sufficiently late in the repeat to permit the mutant DSPP to be nonfunctional but still soluble, will also be recessive in nature and will therefore be found in the very rare patients with recessive dentin diseases. Sequencing analysis of the entire DSPP repeat domain in control samples identified an extraordinary number of SNPs and indels. Within the 30-most 1.2 kb (DSPP HVRR), there were 37 different haplotypes (comprised of specific combinations of 20 indels and 37 SNPs) in 94 individuals; such variance is not commonly seen in coding regions. This high degree of variation may be informative for studies of human migration patterns, including the generally accepted out-of-Africa model. Because of a relatively high rate of change and lack of recombination, mitochondrial and Y chromosome DNA are commonly used to track human geographical lineages even though the information is limited to maternal and paternal lines, respectively. Most autosomal DNA can provide information from both parental lineages; however, interpreting differences among individuals can be complicated by low rates of change (relative to recent human migration events) and by recombination events. DSPP HVRR may be a novel source of information from both parental lineages because it is highly polymorphic and, from our limited sample size, does not appear to be prone to recombination. Thus, DSPP HVRR may provide a valuable tool to supplement mitochondrial and Y chromosome DNA for human migration studies.
Because the large number of indels and SNPs were observed within a small group of humans and because Homo sapiens are generally considered to be genetically narrow due to recent population bottlenecks, we looked further into possible mechanisms. The indels can easily be attributed to the process of slipreplication first proposed by Streisinger et al. [1966] and frequently seen in short DNA repeat structures such as microsatellites. While there is logically a requirement for the template (in deletion formation) or growing DNA replication strand (in insertion formation) to rebind to a perfect (short) or near-perfect (longer) repeat during a replication pause to result in a new indel, the repeats need not be in tandem. Indeed, the various HVRR haplotypes in our small cohort often had indels 49 bp, including one of 189 bp, but always in multiples of nine. The majority of SNPs found in the 37 haplotypes of DSPP HVRR can be attributed to C-to-T transitions of CpG and CpNpG motifs by methylation/deamination. Tabulation of the changes in each nucleotide position in all 204 nominal 9-bp repeat units (AGC AGC GAC) in the full repeat of a representative haplotype (Hap ]1), indicated that the majority of changes were consistent with deamination transitions of the cytosines in CpG and CpNpG motifs. As expected, most changes occurred in the nonomer’s sense strand CpG (64%); although, interestingly, a single CpNpG (CAG) appeared to undergo transitions more often (35%) than even the antisense CpG (16%). Bisulfite sequence analysis of a portion of the DSPP HVRR repeat under stringent conditions verified that the CpApG sense strand sites were sometimes methylated in both ES cell and somatic cell genomic DNA. The cytosines in a CpG unit are well documented to be frequently methylated in the genomic DNA of humans [Robertson and Jones, 2000]. While CpNpG methylation has been mainly studied in plants [Grafi et al., 2007], there are reports of this process in animals, particularly on CpApG motifs [Clark et al., 1995; Haines et al., 2001; Ramsahoye et al., 2000; White et al., 2002]. Ramsahoye et al. [2000] have shown that mouse embryonic stem cells have non-CpG (CpA) methylation due to DNA methyltransferase 3a enzymatic activity. There is a report of CpNpG methylation in human carcinoma [Kouidou et al., 2005] but to our knowledge, ours is the first report of CpNpG methylation in normal human genomic DNA analyzed under conditions sufficiently stringent to fully eliminate incomplete bisulfite reactions. The observed high rate of change in the DSPP repeat’s CpG and CpApG is also consistent with the methylation of these sites in the human germline. Indeed, the large number of SNPs observed within the repeat portion of the DSPP gene in the generally agreed upon genetically narrow human population may suggest that the same locally displaced DNA loop structures that result in the many indels during meiosis may also result in similar structures within this gene at other times. These hairpin loops may cause some portions of the repeat to be held in a partial single-stranded state, thereby increasing the rate of C-to-T deamination in both methylated and nonmethylated cytosines [Lindahl and Nyberg, 1974]. Methylation of mammalian DNA has been hypothesized to occur preferentially on repetitive sequences [Santos et al., 2005], consistent with our observation of the high rate of change of both CpG and CpApG motifs within the DSPP repeat domain.
This study brings the total number of mutations described in DSPP to 14 separate events. The findings here further highlight the phenotypic continuum associated with DSPP mutations, supporting the designation DSPP-associated dentin defects for both DD (II) and DGI (II and III) [Hart and Hart, 2007]. Frameshift mutations in the 9-bp tandem coding repeat of the DSPP gene may turn out to be one of the most frequent causes of DSPP-associated dentin defects. Presumably mutations in the repeat domain of DSPP will be found in many patients linked to the 4q22.1 region in whom mutations are not found outside of the repeat domain [Beattie et al., 2006; Malmgren et al., 2004]. At this time, as shown in Figure 4C, it appears that there are three major consequences of DSPP mutations: alterations of the signal peptide itself; mutations that destroy the conserved IPV domain; and mutations that change the hydrophilic repeat domain to a hydrophobic repeat. This repeat domain has also turned out to be an interesting model system for: 1) the study of indel formation of short tandem repeats larger than what is typically seen in humans; 2) SNP formation due to methylated CpG and CpApG motifs that may occasionally remain in a loop-stabilized single-stranded state subject to unusually rapid deamination transitions; and 3) a rapidly changing autosomal gene useful in the study of the relatedness of human populations around the world.
Supplementary Material
Acknowledgments
We thank Lawrence C. Brody (NHGRI/NIH), Tyra Wolfsberg (NHGRI/NIH), Sarah Tishkoff (University of Maryland), and Karl J. Fryxell (George Mason University) for their many insightful discussions.
Contributor Information
Dianalee A. McKnight, Craniofacial and Skeletal Diseases Branch, NIDCR, NIH, DHHS, Bethesda MD 20892 USA
P. Suzanne Hart, Office of the Clinical Director, NHGRI, NIH, DHHS, Bethesda MD 20892 USA.
Thomas C. Hart, Section of Dental and Craniofacial Genetics, NIDCR, NIH, DHHS, Bethesda MD 20892 USA
James K. Hartsfield, Department of Orthodontics and Oral Facial Genetics, Indiana University School of Dentistry, Indianapolis, IN 46202 USA
Anne Wilson, Department of Pediatric Dentistry, University of Colorado School of Dental Medicine, Aurora, CO 80045 USA.
J. Timothy Wright, Department of Pediatric Dentistry, School of Dentistry, The University of North Carolina, Chapel Hill NC 27599 USA.
Larry W. Fisher, Craniofacial and Skeletal Diseases Branch, NIDCR, NIH, DHHS, Bethesda MD 20892 USA
References
- Ball SP, Cook PJ, Mars M, Buckton KE. Linkage between dentinogenesis imperfecta and Gc. Ann Hum Genet. 1982;46:35–40. doi: 10.1111/j.1469-1809.1982.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Beattie ML, Kim JW, Gong SG, Murdoch-Kinch CA, Simmer JP, Hu JC. Phenotypic variation in dentinogenesis imperfecta/dentin dysplasia linked to 4q21. J Dent Res. 2006;85:329–333. doi: 10.1177/154405910608500409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman JA, Halloran SL, Roulston D, Schwartz S, Suzuki JB, Weitkamp LR, Wenk RE, Wooten R, Cohen MM. An autosomal-dominant form of juvenile periodontitis: its localization to chromosome 4 and linkage to dentinogenesis imperfecta and Gc. J Craniofac Genet Dev Biol. 1986;6:341–350. [PubMed] [Google Scholar]
- Byers PH, Cole WG. Osteogenesis Imperfecta. In: Royce PM, Steinmann BU, editors. Connective tissue and its heritable disorders. 2. New York: Wiley-Liss; 2002. pp. 385–430. [Google Scholar]
- Clark SJ, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- Dean JA, Hartsfield JK, Jr, Wright JT, Hart TC. Dentin dysplasia, type II linkage to chromosome 4q. J Craniofac Genet Dev Biol. 1997;17:172–177. [PubMed] [Google Scholar]
- Delahunty M, Bonifacino JS. Disorders of intracellular protein trafficking in human disease. Connect Tissue Res. 1995;31:283–286. doi: 10.3109/03008209509010824. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- Dong J, Gu T, Jeffords L, MacDougall M. Dentin phosphoprotein compound mutation in dentin sialophosphoprotein causes dentinogenesis imperfecta type III. Am J Med Genet A. 2005;132:305–309. doi: 10.1002/ajmg.a.30460. [DOI] [PubMed] [Google Scholar]
- Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;44(Suppl 1):33–40. [PubMed] [Google Scholar]
- Gajko-Galicka A. Mutations in type I collagen genes resulting in osteogenesis imperfecta in humans. Acta Biochim Pol. 2002;49:433–441. [PubMed] [Google Scholar]
- George A, Srinivasan RSR, Liu K, Veis A. Rat dentin matrix protein 3 is a compound protein of rat dentin sialoprotein and phosphophoryn. Connect Tissue Res. 1999;40:49–57. doi: 10.3109/03008209909005277. [DOI] [PubMed] [Google Scholar]
- Grafi G, Zemach A, Pitto L. Methyl-CpG-binding domain (MBD) proteins in plants. Biochim Biophys Acta. 2007;1769:287–294. doi: 10.1016/j.bbaexp.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Haines TR, Rodenhiser DI, Ainsworth PJ. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–598. doi: 10.1006/dbio.2001.0504. [DOI] [PubMed] [Google Scholar]
- Hart PS, Wright JT, Savage M, Kang G, Bensen JT, Gorry MC, Hart TC. Exclusion of candidate genes in two families with autosomal dominant hypocalcified amelogenesis imperfecta. Eur J Oral Sci. 2003;111:326–331. doi: 10.1034/j.1600-0722.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- Hart PS, Hart TC. Disorders of human dentin. Cells Tissues Organs. 2007;186:70–77. doi: 10.1159/000102682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holappa H, Nieminen P, Tolva L, Lukinmaa PL, Alaluusua S. Splicing site mutations in dentin sialophosphoprotein causing dentinogenesis imperfecta type II. Eur J Oral Sci. 2006;114:381–384. doi: 10.1111/j.1600-0722.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Nam SH, Jang KT, Lee SH, Kim CC, Hahn SH, Hu JC, Simmer JP. A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum Genet. 2004;115:248–254. doi: 10.1007/s00439-004-1143-5. [DOI] [PubMed] [Google Scholar]
- Kim JW, Hu JC, Lee JI, Moon SK, Kim YJ, Jang KT, Lee SH, Kim CC, Hahn SH, Simmer JP. Mutational hot spot in the DSPP gene causing dentinogenesis imperfecta type II. Hum Genet. 2005;116:186–191. doi: 10.1007/s00439-004-1223-6. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;86:392–399. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- Kouidou S, Agidou T, Kyrkou A, Andreou A, Katopodi T, Georgiou E, Krikelis D, Dimitriadou A, Spanos P, Tsilikas C, Destouni H, Tzimagiorgis G. Non-CpG cytosine methylation of p53 exon 5 in non-small cell lung carcinoma. Lung Cancer. 2005;50:299–307. doi: 10.1016/j.lungcan.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Gu TT, DuPont BR. Assignment of dentin sialophosphoprotein (DSPP) to the critical DGI2 locus on human chromosome 4 band q21.3 by in situ hybridization. Cytogenet Cell Genet. 1997a;79:121–122. doi: 10.1159/000134697. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J Biol Chem. 1997b;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- Malmgren B, Lindskog S, Elgadi A, Norgren S. Clinical, histopathologic, and genetic investigation in two large families with dentinogenesis imperfecta type II. Hum Genet. 2004;114:491–498. doi: 10.1007/s00439-004-1084-z. [DOI] [PubMed] [Google Scholar]
- Marini JC. Osteogenesis imperfecta: comprehensive management. Adv Pediatr. 1988;35:391–426. [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative highresolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbureke KU, Fisher LW. Expression of SIBLINGs and their partner MMPs in salivary glands. J Dent Res. 2004;83:664–670. doi: 10.1177/154405910408300902. [DOI] [PubMed] [Google Scholar]
- Ogbureke KU, Fisher LW. Renal expression of SIBLING proteins and their partner matrix metalloproteinases (MMPs) . Kidney Int. 2005;68:155–166. doi: 10.1111/j.1523-1755.2005.00389.x. [DOI] [PubMed] [Google Scholar]
- Ogbureke KU, Fisher LW. Sibling expression patterns in duct epithelia reflect the degree of metabolic activity. J Histochem Cytochem. 2007;55:403–409. doi: 10.1369/jhc.6A7075.2007. [DOI] [PubMed] [Google Scholar]
- Pallos D, Hart PS, Cortelli JR, Vian S, Wright JT, Korkko J, Brunoni D, Hart TC. Novel COL1A1 mutation (G559C) [correction of G599C] associated with mild osteogenesis imperfecta and dentinogenesis imperfecta. Arch Oral Biol. 2001;46:459–470. doi: 10.1016/s0003-9969(00)00130-8. [DOI] [PubMed] [Google Scholar]
- Paulin R, Grigg GW, Davey MW, Piper AA. Urea improves efficiency of bisulphite-mediated sequencing of 50-methylcytosine in genomic DNA. Nucleic Acids Res. 1998;26:5009–5010. doi: 10.1093/nar/26.21.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- Qin C, Brunn JC, Cadena E, Ridall A, Tsujigiwa H, Nagatsuka H, Nagai N, Butler WT. The expression of dentin sialophosphoprotein gene in bone. J Dent Res. 2002;81:392–394. doi: 10.1177/154405910208100607. [DOI] [PubMed] [Google Scholar]
- Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet. 2002;11:2559–2565. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Santos KF, Mazzola TN, Carvalho HF. The prima donna of epigenetics: the regulation of gene expression by DNA methylation. Braz J Med Biol Res. 2005;38:1531–1541. doi: 10.1590/s0100-879x2005001000010. [DOI] [PubMed] [Google Scholar]
- Shields ED, Bixler D, el-Kafrawy AM. A proposed classification for heritable human dentine defects with a description of a new entity. Arch Oral Biol. 1973;18:543–553. doi: 10.1016/0003-9969(73)90075-7. [DOI] [PubMed] [Google Scholar]
- Song Y, Wang C, Peng B, Ye X, Zhao G, Fan M, Fu Q, Bian Z. Phenotypes and genotypes in 2 DGI families with different DSPPmutations. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:360–374. doi: 10.1016/j.tripleo.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;278:24874–24880. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- Streinsinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO-T cells. J Immunol. 2002;168:2820–2827. doi: 10.4049/jimmunol.168.6.2820. [DOI] [PubMed] [Google Scholar]
- Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, Wu G, Qiang B, Lo WH, Shen Y. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet. 2001;27:151–152. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen L, Liu J, Zhao Z, Qu E, Wang X, Chang W, Xu C, Wang QK, Liu M. A novel DSPP mutation is associated with type II dentinogenesis imperfecta in a Chinese family. BMC Med Genet. 2007;8:52. doi: 10.1186/1471-2350-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.