Figure 5. Pro-Code/CRISPR screen for genes conferring sensitivity or resistance to antigen-dependent T cell killing.
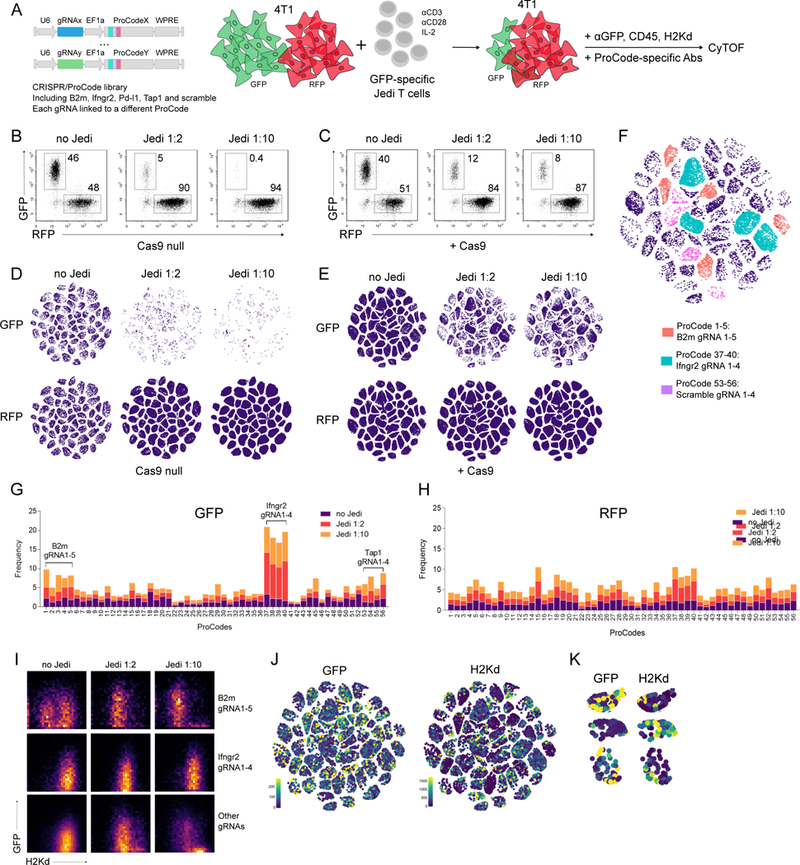
(A) Schematic of the immune editing co-culture system and the ProCode/CRISPR library. 4T1 cells (+/−Cas9, +/−GFP/RFP) were transduced with a library of 56 ProCode/CRISPR vectors, co-cultured with activated Jedi T cells, and analyzed by CyTOF. (B) Frequency of GFP+ and RFP+ 4T1 cells was measured by flow cytometry. Representative dotplots are shown. Jedi 1:2 and Jedi 1:10 is 2-fold and 10-fold multiple of T cells to cancer cells, respectively. (C) Frequency of GFP+ and RFP+ 4T1-Cas9 cells was measured by flow cytometry. Representative dotplots are shown. (D) viSNE visualization of the 4T1-GFP and 4T1-RFP Pro-Code populations co-cultured alone or with activated Jedi T cells. Each cluster corresponds to a different Pro-Code. (E) viSNE visualization of the 4T1-GFP-Cas9 and 4T1-RFP-Cas9 Pro-Code populations co-cultured alone or with activated Jedi T cells. Each cluster corresponds to a different Pro-Code. (F) viSNE visualization of 56 Pro-Code/CRISPR populations (GFP-4T1-Cas9, Jedi 1:10) colored by the target: orange=B2m, cyan=Ifngr2, purple=scramble, navy=others. (G, H) Frequency of each Pro-Code/CRISPR populations among the GFP-4T1-Cas9 (G) and RFP-4T1-Cas9 (H) cells in the absence (no Jedi) or presence (Jedi 1:2, Jedi 1:10) of GFP- specific Jedi T cells. (I) GFP and H2Kd expression on 4T1-Cas9-GFP cells expressing gRNAs targeting B2m, Ifngr2 and all other genes. (J) GFP and H2Kd expression levels Pro-Code/CRISPR populations in GFP-4T1-Cas9 cells resisting T cell killing (Jedi 1:10); (K) GFP and H2Kd expression on selected Pro-Code populations (from J). Data is representative of 3 independent experiments.