Abstract
microRNAs (miRNAs) are generated from long primary (pri-) RNA polymerase II (Pol II)–derived transcripts by two RNase III processing reactions: Drosha cleavage of nuclear pri-miRNAs and Dicer cleavage of cytoplasmic pre-miRNAs. Here we show that Drosha cleavage occurs during transcription acting on both independently transcribed and intron-encoded miRNAs. We also show that both 5′-3′ and 3′-5′ exonucleases associate with the sites where co-transcriptional Drosha cleavage occurs, promoting intron degradation before splicing. We finally demonstrate that miRNAs can also derive from 3′ flanking transcripts of Pol II genes. Our results demonstrate that multiple miRNA-containing transcripts are co-transcriptionally cleaved during their synthesis and suggest that exonucleolytic degradation from Drosha cleavage sites in pre-mRNAs may influence the splicing and maturation of numerous mRNAs.
miRNAs belong to a growing class of conserved noncoding transcripts, 21–23 nucleotides (nt) long, that regulate the fine tuning of gene expression by RNA-mediated gene-silencing mechanisms1. They derive from a longer primary transcript, the pri-miRNA, by a stepwise process that occurs in both nuclear and cytoplasmic compartments. In the nucleus, the Microprocessor complex containing Drosha, an RNase III like enzyme, and its cofactor DGCR8 generates a pre-miRNA hairpin product about 70 nt long2–4. Pri-miRNA processing represents a crucial step in miRNA biogenesis because it defines the miRNA sequences embedded in long pri-miRNAs and generates one end of the mature miRNA molecule. DGCR8 is thought to recognize the junction between the single- and double-stranded region (SD junction) and the 33-base pair (bp) stem of the pri-miRNA substrate. The double-stranded RNA (dsRNA) binding domain (dsRBD) of Drosha may then transiently interact with the stem, allowing the catalytic center of the enzyme to cleave the RNA duplex about 11 bp from the SD junction5. The receptor Exportin 5 exports the pre-miRNA products to the cytoplasm6, where a second RNase III-like enzyme, Dicer, converts them into mature miRNA duplexes. Last, following strand selection and separation of the duplex, the mature 22-nt miRNA is incorporated into the RNA-induced silencing effector complex (RISC). This complex targets specific mRNAs, resulting in their inactivation through either degradation or translational repression7–9.
Analysis of the genomic localization of human miRNA sequences reveals that they are present either as part of independent Pol II transcription units or within annotated ‘host’ genes10–12. Approximately 50% of human miRNAs are thought to be expressed from gene introns13,14. The expression pattern of host gene transcripts and their miRNAs suggests that mammalian miRNAs are transcriptionally linked to the expression of their host genes13,15. In addition, several chimeric transcripts containing miRNA sequences and part of the adjacent mRNA sequence have been found from expressed sequence tag (EST) analyses16. Notably, some of these EST fragments are partially spliced, with either 5′ or 3′ ends that match putative Drosha cleavage sites14. A final link between pre-mRNA and intronic miRNA processing is derived from evidence that miRNA-encoding introns are spliced more slowly than adjacent introns. This suggests that Microprocessor binding somehow interferes with the splicing reaction14.
The above evidence suggests that maturation of intronic miRNAs may be closely associated with intronic splicing, consistent with the mechanism of intronic small nucleolar RNA (snoRNA) maturation17,18. Here physical and functional links between intronic snoRNP assembly, pre-mRNA synthesis and processing have also been described19–21. We demonstrate in human HeLa cells that pre-miRNAs are generated through co-transcriptional cleavage by Drosha. This occurs both in independently transcribed (intergenic) miRNA genes and from introns harboring miRNA sequences. Furthermore, the Microprocessor complex as well as 5′-3′and 3′-5′ RNA exonucleases are recruited to chromatin associated with intronic miRNAs during transcription of the host primary transcript, and Drosha cleavage occurs before host intron splicing. We also show that the miRNA hairpin does not affect the accumulation of the host-gene mRNA, predicting that mature miRNAs and mRNAs derive from a common nascent transcript.
Results
Co-transcriptional recruitment of Drosha
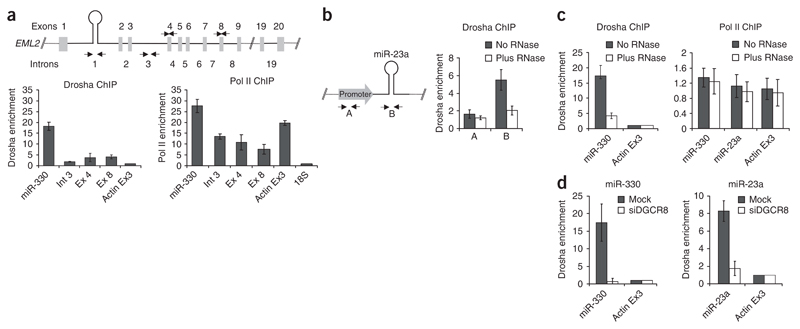
We carried out chromatin immunoprecipitation (ChIP) assays using a Drosha-specific antibody on HeLa cell chromatin of endogenous, intergenic or intron-encoded miRNA genes. Intergenic miR-7, miR-223, miR-23a and let-7a-1 miRNA genes and two host genes, the echinoderm microtubule-associated protein–like 2 (EML2) and minichromosome-maintenance complex component 7 (MCM7) genes, which contain miR-330 and miR-25 located in the first intron of EML2 and the thirteenth intron of MCM7, respectively, were tested (Fig. 1). Drosha specifically associates with chromatin of the miR-23a and let-7a1 genes (Fig. 1a, Drosha ChIP) but not with chromatin of the miR-223 and mir-7 genes, which are not expressed in HeLa cells22,23. Similarly, Drosha recruitment was also observed on both intronic miRNA loci (Fig. 1b, Drosha ChIP) but not on GAPDH and actin genes, used as negative controls (Fig. 1a,b). We tested the expression profile of all miRNA loci and that of GAPDH and actin genes using Pol II immunoprecipitation (Fig. 1a,b, Pol II ChIP). Notably, the binding of Drosha correlates with the presence of Pol II on the same miRNA loci, suggesting that its recruitment is associated with transcription. We also analyzed the chromatin association of Drosha for the tissue-specific miR-223 gene, a known modulator of human myeloid differentiation, which is activated during retinoic acid–induced differentiation of NB4 cells22 (Fig. 1c, northern blot). ChIP analysis showed that the recruitment of Drosha on miR-223 in NB4 cells (Drosha ChIP) follows its activation (Pol II ChIP), establishing that Drosha recruitment requires ongoing transcription of the miR-223 gene (Fig. 1c).
Figure 1. Chromatin association of the Microprocessor complex.
(a) Association of Drosha and Pol II with miRNA gene chromatin in HeLa cells. The diagram depicts the miRNA gene (‘//’ denotes chromosomal integration), with ChIP analysis using either Drosha (left) or Pol II (right)–specific antibodies presented below. Results are based on quantitative real-time PCR analysis (Methods) using four different miRNA-specific probes and negative control probes for GAPDH and tRNA genes. Error bars show s.e.m. based on three independent experiments. (b) ChIP analysis of two endogenous intronic miR-330 and miR-25 miRNA sequences using Drosha and Pol II antibodies in HeLa cells. Maps of host EML2 and MCM7 genes are shown above the quantitative ChIP analysis. (c) Analysis of co-transcriptional recruitment of Drosha to the endogenous miR-223 gene following transcriptional activation by retinoic acid (RA) in NB4 cells. Northern blot analysis shows the progressive appearance of miR-223 following RA treatment. U2 snRNA was used as a positive control. This profile is consistent with increasing ChIP signals for Pol II and Drosha chromatin association. (d) miR-223 pcDNA plasmid constructs driven by the miR-23a promoter with (WT) or without (Δ) the 5′ single-stranded region of miR-223 (left) were transfected into HeLa cells, and total RNA was analyzed by northern blot (middle) or chromatin by Drosha ChIP (right). U2 and miR-23a probes provide positive controls.
Next, we generated two different constructs in which the pri–miR-223 coding sequence, with (wild type) or without (Δ) the SD junction required for efficient Drosha cleavage5, was positioned downstream of the ubiquitous miR-23a~27a~24-2 promoter24. After transfecting the constructs into HeLa cells, we isolated the total RNA and analyzed the nuclear chromatin (Fig. 1d). The wild-type but not the Δ construct generated mature miR-223 and yielded a positive Drosha ChIP signal. As positive controls, U2 small nuclear RNA (snRNA) for the northern blot and endogenous miR-23a for the Drosha ChIP gave similar signals in both wild-type and Δ samples (Fig. 1d).
To validate the specificity of Microprocessor complex recruitment, we performed ChIP analysis across the host EML2 gene and used oligonucleotide pairs to detect the promoter and coding region of the miR-23a gene (Fig. 2a,b). Notably, we observed only Drosha over the miR-330 and miR-23a sequences. Acting as a further control, we detected Pol II ChIP signals across the EML2 gene, even though a two-fold higher signal was observed nearer the 5′ end, presumably related to promoter-proximal pausing25. We furthermore determined whether the interaction of the Microprocessor with chromatin occurs through association with the nascent transcript by testing the effect of RNase treatment before the immunoprecipitation step in the ChIP analysis. These results (Fig. 2b,c) show that Drosha association with the chromatin both at miR-23a and miR-330 sequences is RNase sensitive, whereas the Pol II association is unaffected (Fig. 2c, right). Previous in vitro analysis has shown that the Microprocessor complex interacts with pri-miRNAs through DGCR8, which possesses tandem dsRBDs5,26. We therefore tested whether Drosha association with mir-23a and mir-330–containing chromatin depends on DGCR8 by depleting DGCR8 levels using RNA interference (RNAi). miR-23a processing is indeed impaired (Supplementary Fig. 1 online), and we also observed a substantial reduction in Drosha levels at the miRNA loci tested (Fig. 2d). These results suggest that, in vivo, DGCR8 may bind pri-miRNA hairpins before Drosha.
Figure 2. Specificity of the Drosha–miRNA chromatin interaction.
(a) Diagram of the EML2 gene indicating the positions of exons, introns and the miR-330 hairpin. Arrows demonstrate positions of the PCR primers used to determine the specificity of the Drosha ChIP signal. ChIP analysis below (as in Figure 1) shows the enrichment of the Drosha ChIP signal over intron 1, whereas Pol II signals are observed for all probed regions. (b) The miR-23a Drosha ChIP signal (probe B) which also shows RNase sensitivity compared to the promoter region (probe A). (c) miR-330 Drosha ChIP signal shows RNAse sensitivity (with or without RNAse) compared to the actin control. For both miR-23a and miR-330, the Pol II ChIP signals, in contrast to the Drosha ChIP signals, are unaffected by RNase treatment. (d) Drosha ChIP analysis on endogenous miR-330 and miR-23a using HeLa cells depleted for DGCR8 by RNAi. Histograms show ChIP results based on quantitative real-time PCR analysis and demonstrate that Drosha recruitment is DGCR8 dependent.
Microprocessor cleaves pre-miRNA harpin before splicing
We previously showed that co-transcriptional cleavage of nascent intronic transcripts does not affect the efficiency of splicing, predicting that exons of pre-mRNA are tethered to the elongating RNA Pol II either directly or indirectly27,28. Our results demonstrate that the Microprocessor complex is co-transcriptionally recruited to chromatin of Pol II host miRNA genes (Figs. 1 and 2), and for this reason, we predict that the pre-miRNA sequence is excised from introns during transcription without affecting the splicing of the host gene.
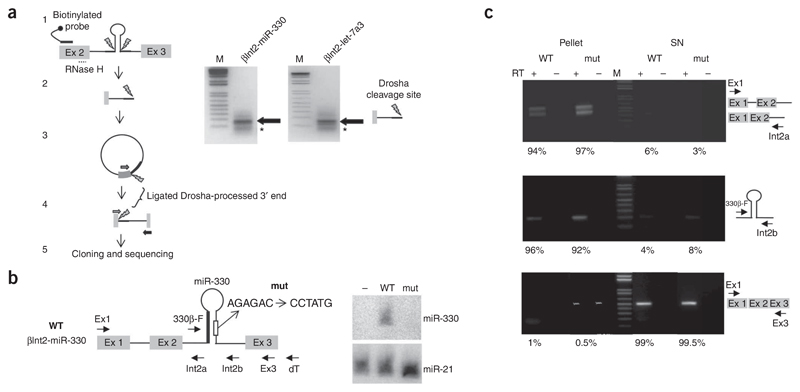
We generated two constructs, βInt2-miR-330 and βInt2-miR-let-7a3, containing miR-330 and let-7a3 pre-miRNA hairpins in the middle of the second intron of the β-globin gene transcribed from the HIV-1 promoter (Fig. 3a). After transfection into HeLa cells, we carried out northern blotting of the cytoplasmic RNA. Both miR-330 and let-7a3 mature miRNAs were readily detected by northern blot analysis, indicating their efficient processing and proper localization to the cytoplasmic fraction (Fig. 3a, right). Note that in HeLa cells, endogenous miR-330 is expressed at low levels (Supplementary Fig. 2a online), even though the miR-330 host gene EML2 is expressed in HeLa cells14 and both Pol II and Drosha associate with the miR-330–containing intron 1 (Fig. 1b). It is possible that down-regulation of miR-330 processing occurs as described for other miRNA-containing genes29,30.
Figure 3. miRNA maturation from the second intron and 3′ flanking region of the β-globin gene in HeLa cells.
(a) Diagram of the β-globin construct with or without insertion of either miR-330 or let-7a3 pre-miRNAs in intron 2 (βInt2-miR-330 or βInt2-let-7a3 constructs). The exons (gray boxes), introns (lines) and miRNAs (white box). Below, northern blot analysis of cytoplasmic RNA from transfected HeLa cells. Specific antisense oligonucleotides were used to detect miR-330, let-7a3 and miR-21 miRNAs. Lower northern blots detect endogenous miR-21, used as a loading control. (b) Diagram of βInt2-miR-330 showing positions of the NRO probes (underlined). ‘M’ denotes background signal. The HIV-1 promoter with the 5′ portion of the β-globin gene as a dashed line and the backbone plasmid are indicated as is the position of a biotinylated probe (Bio-int2). Graph shows the ratio between B3 and a hybridization signal (left) from selected and unselected fractions. (c) β-globin gene constructs with 3′ flanking CoTC and miRNA sequences shown as white boxes and CoTC deletion indicated by a white triangle. NRO probe positions are underlined. Left, NRO analysis of HeLa cells transiently transfected with constructs indicated. Whereas the β construct shows dramatic Pol II termination after probe B4, ΔCoTC and β3′-miR-330 and β3′-let-7a3 demonstrate read-through signals around the transfected plasmids, as detected by high signals over probes A and U3. Right, northern blot analysis of miRNAs produced from the indicated plasmid constructs, indicating that 3′ flanking and intronically located pre-miRNAs are expressed at similar levels.
The possibility that these intronically positioned miRNA sequences are co-transcriptionally cleaved was analyzed by a refinement of the nuclear run-on (NRO) protocol previously used to characterize intron co-transcriptional cleavage events27. We transiently transfected HeLa cells with βInt2-miR-330, isolated the cell nuclei and subjected them to run-on analysis. An antisense biotinylated probe complementary to the intronic sequence, upstream of the Drosha cleavage site was then used to select these nascent transcripts (Fig. 3b, diagram) and the selected and unselected fractions were hybridized to separate filters containing single-stranded M13 DNA probes (Fig. 3b, Select and Unselect). In parallel, as a control, we carried out a regular NRO experiment on the same construct, and this produced strong hybridization signals over all gene probes (Fig. 3b, NRO). The selected and unselected fractions showed differences in hybridization signal beyond probe a, located upstream of the Drosha cleavage site (B3/a ratio, Fig. 3b, histogram). Thus, the unselected fraction gave a higher B3/a value than the selected fraction. These results suggest that co-transcriptional cleavage of the nascent transcript occurs over the pre-miRNA sequence and consequently confirms that Drosha cleavage is a co-transcriptional event. However, as there is partial selection of transcripts upstream of the Drosha cleavage site, these data also indicate that Drosha cleavage is a relatively slow process.
To compare Drosha cleavage with another well-characterized co-transcriptional cleavage event, we cloned miR-330 and let-7a3 hairpins downstream of the poly(A) site of the β-globin gene in place of its co-transcriptional cleavage (CoTC) termination sequence (Fig. 3c, diagram). Transcriptional termination relies on the specific CoTC sequence, which is cleaved co-transcriptionally, resulting in the recruitment of the nuclear 5′-3′ exonuclease XRN2 (ref. 31). We carried out NRO analysis (NRO, Fig. 3c, left) on these constructs using single-stranded M13 probes spanning the β-globin gene and an M13 (M) alone control. Whereas lower hybridization signals were observed over the probes A and U3 in the β-globin construct owing to efficient transcriptional termination, stronger signals were observed for the termination-deficient ΔCoTC construct and those containing miRNA hairpins in the 3′ region. This suggests that Drosha co-transcriptional cleavage does not promote efficient transcription termination. Together, these results (Fig. 3b,c) suggest that Drosha pre-miRNA processing, although a co-transcriptional mechanism, is kinetically slower than CoTC cleavage and so is unable to promote efficient transcriptional termination downstream of the β-globin poly(A) site.
We next determined whether pre-miRNA maturation occurs from β3′-miR-330 and β3′-let-7a3 gene constructs by northern blot analysis (Fig. 3c, right). We observed accumulation of mature miRNAs from both constructs at comparable levels to the intronic constructs βInt2-miR-330 and βInt2-let-7a3. These results demonstrate that pre-miRNAs may be processed from the 3′ flanking regions of a gene if extended 3′ transcription occurs. As these transcripts are highly unstable and remain tethered to the elongating Pol II complex32, this confirms that pre-miRNA maturation occurs co-transcriptionally.
We next used hybrid selection circular rapid amplification of cDNA ends (hscRACE33) to establish that Drosha cleavage occurs on nascent transcripts. βInt2-miR-330 and βInt2-let-7a3 nuclear RNA were hybridized to a biotinylated antisense RNA probe complementary to the second exon of the β-globin gene to select nascent transcripts. The hscRACE procedure is described in Figure 4a. For both selected RNA samples, we observed one major hscRACE DNA product, which, based on sequence analysis (data not shown), corresponds to unspliced RNA up to the Drosha cleavage site; we also observed some minor bands that probably correspond to degradation intermediates (Fig. 4a). We also included controls to show that the hscRACE products correspond to in vivo Drosha cleavage products rather than nonspecific RNAs generated by the hscRACE technique (Supplementary Fig. 2b,c)
Figure 4. Co-transcriptional processing of pre-miRNAs from the β-globin gene intron 2.
(a) hscRACE procedure31,55 A diagram of β-globin transcript containing the pre-miRNA in intron 2 is shown (1). Grey boxes are exons whereas miRNA sequence is indicated by a hairpin within intron 2 (solid line). Drosha cleavage sites are shown as lightening bolts. Hybrid selection of this transcript was carried out using antisense biotinylated RNA (black line and circle) and selected transcripts were released by RNase H digestion directed by antisense DNA oligonucleotide (dotted line). Released RNAs were circularized by RNA ligation and reverse transcribed with a primer (arrow) across the ligation junction (2,3). PCR amplification using a primer pair (gray and black arrows) amplifies only cDNA obtained from the ligated RNA (4). Products were analyzed on agarose gel, cloned and sequenced (5). Right, agarose gel analysis of hscRACE products obtained from βInt2-miR-330 and βInt2-miR-let7a3 constructs. Major products are indicated by arrows and minor products by an asterisk. M indicates the molecular weight DNA marker. (b) Diagram of chimeric βInt2-miR-330 transcript with labeled arrows indicating the primers used for RT-PCR analysis. Hairpin sequences depict the miR-330 stem-loop with the mature miRNA position sequence as a thick line and the mutant sequence change as indicated. Right, northern blot analysis of miRNAs produced from the wild-type and mut plasmid constructs. Below, an endogenous miR-21 control. (c) RT-PCR analysis of the chromatin-associated (Pellet) and nucleoplasmic (SN) fractions carried out with the primer pairs indicated to the right of the agarose gels. Identities of the PCR products are shown to the right. Quantitative analysis was carried out by real-time PCR, as shown below as % RNA in Pellet or SN fractions.
Overall, these data confirm that the RNA 3′ ends observed by hscRACE analysis were authentic in vivo products of Drosha cleavage. In addition, sequence analysis of the main PCR products revealed that the 3′ processed molecules still contained the exon 2–intron 2 junction region. Consequently, we provide direct evidence that unspliced pre-mRNA transcripts are major substrates of Drosha processing.
Co-transcriptional Drosha cleavage of intronic microRNAs
Our above results demonstrate that the Microprocessor complex binds miRNA sequences during transcription and that Drosha processing occurs before the host intron is spliced out. However, to determine whether nascent RNA, rather than an unprocessed, released transcript, is the actual substrate for Drosha cleavage, we used a fractionation procedure to separate chromatin-associated nuclear RNA from released nucleoplasmic transcripts27,34.
We transfected HeLa cells with the βInt2-miR-330 plasmid or a mutant version containing altered sequences in the miR-330 stem region that are known to strongly inhibit Drosha processing14. We confirmed that mature miRNA was not detectable by northern blot analysis for mutant miR-330 (Fig. 4b). Following transfection, equal amounts of RNA extracted from chromatin-associated (Pellet) and nucleoplasmic fractions were subjected to reverse-transcription PCR (RT-PCR) analysis to detect the distribution of β-globin transcripts containing unspliced introns 1 and 2 (primers Ex1-Int2a and 330β-F-Int2b). Intron 1 and/or the miRNA harboring intron 2 were predominantly localized in the chromatin-associated (greater than 90%) as compared to the nucleoplasmic fraction (Fig. 4c, above and middle, Pellet and SN). Note that all these RT-PCR data were also analyzed by quantitative real-time PCR analysis, as shown below the gel images. Little β-globin transcript was released from the site of transcription before all the introns were spliced out. Both the wild-type and mutant constructs showed similar distributions, indicating that a functional intronic miRNA does not change the chromatin-nucleoplasmic localization of β-globin transcripts. Note that a higher pellet signal was observed for mutant RNA across intron 2 (Fig. 4c, middle), presumably because of the lack of Drosha cleavage. As a control, the same RNA samples were reverse transcribed in the presence of oligodT, and the cDNA was amplified using Ex1 and Ex3 PCR primers (Fig. 4c, below). A band corresponding to the processed β-globin mRNA was present in only the supernatant fraction (SN), showing that spliced transcripts are readily detected in this fraction. As miRNA-harboring transcripts are preferentially detected in the chromatin-associated fraction, both pre-miRNA cleavage and intronic splicing must occur on the same nascent transcripts.
Drosha cleavage promotes intron degradation by exonucleases
We reasoned that co-transcriptional cleavage of intronic miRNAs by Drosha might be associated with recruitment of 5′-3′ and 3′-5′ exonucleases to the newly generated 5′ and 3′ ends of the host intron transcript. Therefore, we performed ChIP analysis of the EML2 and MCM7 genes using antibodies against both the XRN2 5′-3′ exonuclease (human homolog of the yeast Rat1) and the PMscl100 component of the nuclear Exosome (Fig. 5a,b). When compared to the actin gene, used as a negative control, enrichment was observed for XRN2 exonuclease on the intronic regions containing miR-330 and miR-25 (Fig. 5b). Similarly, an enrichment of PMscl100 was detected over both introns containing miRNA sequences as compared to other gene sequences (Fig. 5b). Co-localization of the XRN2 and Exosome exonuclease activities as well as Drosha over host introns containing miRNA sequences strongly supports the co-transcriptional miRNA-processing model and further suggests that after Drosha cleavage the intronic sequences are co-transcriptionally degraded.
Figure 5. Exonuclease activities are associated with chromatin of intronic pre-miRNAs.
(a) Schematic representation of two miRNA-hosting genes EML2 and MCM7, demonstrating positions of pre-miRNAs (hairpins) and PCR primer locations. (b) Real-time ChIP analysis using XRN2 and PMscl100 antibodies with the indicated PCR primers. (c) Diagram shows the position of exons (gray boxes), introns (solid lines) and primers (arrows) used to detect unspliced and spliced transcripts of the MCM7 and CTDSPL genes. Graphs show results from quantitative real-time RT-PCR analysis performed on total RNA extracted from HeLa cells treated with siRNAs targeting XRN2, PMscl100 and Drosha mRNAs. (d) Genomic location of miRNAs relative to their hosting genes. miRNAs in introns and exons are separated according to the nature of the hosting gene (protein-coding versus noncoding (nc) ESTs). miRNAs in alternatively spliced transcripts (Protein coding—between shuffled exons) as well as the intergenic regions are also presented in the table.
To determine the effect of both exonucleases and Drosha on the splicing efficiency of miRNA-containing introns, we performed RNAi-mediated knockdown in HeLa cells of XRN2, Exosome (PMscl100) and Drosha. In each case, protein levels were substantially reduced compared to mock depleted cells (Supplementary Fig. 1a). We then measured the accumulation of unspliced versus spliced forms of endogenous gene transcripts hosting intronic miRNAs, following knockdown of exonucleases. We selected two specific miRNA-hosting genes, MCM7 and CTDSPL, which contain miRNA sequences in a short and a long intron, respectively (Fig. 5c). Total RNA was extracted from both mock and siRNA-treated cells and reverse transcribed with a gene-specific oligonucleotide. Primers complementary to the miRNA-harboring intron and to the neighboring exons were used to quantitatively detect unspliced and spliced transcripts, respectively (Fig. 5c). In both miRNA-hosting genes, the splicing efficiency of the miRNA-containing intron was reduced with either XRN2 or PMscl100 knockdown, regardless of intron length (Fig. 5c, unspliced/spliced ratio), whereas a weaker effect was observed in Drosha-depleted cells. These results confirm that, although co-transcriptional cleavage of the intronic nascent transcript does not affect splicing efficiency27, in introns containing miRNAs, clearance of intronic sequences following Drosha cleavage may act to enhance the splicing efficiency.
Some intronic sequences are known to contain regulatory signals that are important to enhance or prevent the inclusion of an alternative exon in the mature transcript. We therefore reasoned that co-transcriptional degradation of such introns could detrimentally affect the splicing pattern of specific transcripts. We used a bioinformatics approach to analyze the distribution of intronic miRNA hairpins in the human genome, matching the miRNA location with protein-coding genes. Among 344 intronic miRNAs analyzed, 243 (70%) are located within introns of characterized protein genes (miRNA introns, Fig. 5d), and they are furthermore preferentially located near the middle of the intron (data not shown). Notably, recent analysis of intron size in the human and mouse genomes has shown that miRNA-harboring introns are generally larger than other introns, even comparing introns of the same host gene35. Finally, when refining the bioinformatic analysis for alternatively spliced transcripts, we observe that, even though 70–80% of human genes are alternatively spliced36,37, only 2% of intron-encoded miRNAs are located in introns adjacent to alternatively spliced exons. Consequently, our data suggest that during evolution, the presence of co-transcriptionally cleaved sequences within introns and subsequent degradation events may have acted as a driving force for the intron size by excluding the presence of miRNAs near key sites of splicing regulation.
Discussion
It is well established that the major RNA-processing activities acting on pre-mRNAs such as capping, splicing and 3′ end cleavage and polyadenylation occur co-transcriptionally. Furthermore, the recruitment of these activities to Pol II transcription sites is associated either directly or indirectly with the crucial C-terminal domain of the large Pol II subunit38. It is also apparent that mRNA editing performed by the ADAR enzyme takes place on pre-mRNA co-transcriptionally39. Of these RNA-processing activities, only 3′ end cleavage breaks the continuity of the pre-mRNA, allowing accessibility of the resulting ends to exonucleolytic degradation. In particular, the 5′-3′ exonuclease XRN2 promotes Pol II termination by trimming the cut-off product after 3′ end processing40,41. The 5′ product is protected from Exosome 3′-5′ degradation by 3′ polyadenylation and the association of poly(A) binding proteins42.
We now establish that Drosha-mediated cleavage to generate pre-miRNAs also occurs co-transcriptionally, consistent with previous studies14. Thus, in both dedicated miRNA-expressing genes and in host genes with miRNA sequences resident within their introns, Drosha is detected on chromatin associated with these transcribed miRNA sequences. For intronically located pre-miRNAs, our data predict that the host introns will be cleaved before splicing, therefore exposing the intron transcript to rapid exonucleolytic degradation. Thus, for the two intronic pre-miRNAs investigated, both Exosome and XRN2 interact with the chromatin of these specific introns, effectively providing a nuclease signature of associated RNA-processing activities at these gene locations. A related nuclease signature has been described for the introns of two Pol II–transcribed genes, RPS22B and RPL18A, of Saccharomyces cerevisiae43. For these gene pre-mRNAs, hairpin structures present in their single introns are cleaved by the sole RNase III activity in S. cerevisiae, Rnt1. This cleavage site is recognized by both Exosome and Rat1 (XRN2 homolog) and is thought to cause rapid transcript turnover, thereby regulating gene expression. It is apparent that these S. cerevisiae genes have a related mechanism of intronic RNA degradation, although this eukaryote does not produce any miRNAs. Overall, our results suggest that in higher eukaryotes, following Drosha cleavage, the co-transcriptional exonucleolytic clearance of the extraneous intronic sequences may increase splicing efficiency by facilitating focus of the spliceosome on exonic regions.
We previously demonstrated that co-transcriptional cleavage of introns by the artificial insertion of CoTC or hammerhead ribozyme sequences does not impair the efficiency of splicing27. However, in the case of alternatively spliced exons, positioning ribozymes between intronic splicing regulatory sequences of introns can influence the pattern of alternative splicing28. These results are taken to imply that exons may be tethered to the elongating Pol II complex in such a way that exonucleases acting on the exposed intronic RNA ends are restricted from reading into the adjacent exons, which might otherwise ablate exon splicing. Our studies on the co-transcriptional processing of pre-miRNA generalize these previous results by showing that numerous introns are subjected to co-transcriptional cleavage. Previous bioinformatic analysis14 and the data presented here indicate that almost half of miRNAs mature from intronic locations, suggesting that many introns will suffer co-transcriptional cleavage and potential exonucleolytic degradation. Notably, only a limited number of miRNAs are present in introns adjacent to alternatively spliced exons. Moreover, the long intron sizes and the preferential central position of the miRNA hairpin inside the hosting intron may act to ensure that co-transcriptional cleavage and degradation does not affect splicing. Added to this list, the rare examples of intronic snoRNAs may similarly promote intronic cleavage20,44. All of these intronic cleavage events, that release either pre-miRNAs or snoRNAs, will require exon tethering to secure the appropriate splicing of the host gene’s mRNA.
A physical link between pre-miRNA processing and splicing is also supported by evidence that Drosha and DGCR8 associate with several pre-mRNA–processing proteins4,45. In addition, the general RNA binding protein, hnRNP A1, involved in the regulation of alternative splicing has a role in the production of intronic miR-18a46. It is possible that in mammals, Drosha–DGCR8 may exist as multiple complexes with different RNA-processing factors.
As in higher eukaryotes, Pol II transcription often extends up to 1 kb or even further beyond the poly(A) signal; it seems plausible that a hitherto unidentified class of miRNAs may exist at these locations. We show that miRNAs located in the 3′ flanking region of Pol II transcripts can be produced as efficiently as when they are present within an intron. It is possible that some miRNAs, at present assumed to derive from intergenic genes, may in reality come from read-though transcription of upstream Pol II–transcribed genes.
A caveat to the interpretation of our experimental results is that, although we have demonstrated Drosha cleavage occurs co-transcriptionally on chromatin-associated pre-mRNAs, we cannot prove that a single pre-mRNA molecule acts as the precursor for both pre-miRNA and mRNA synthesis. However, our previous studies indicate that exons are tethered to the Pol II elongation complex, providing a clear mechanism to facilitate splicing of discontinuous pre-mRNA sequences as generated by co-transcriptional cleavage of intronic pre-miRNA. This work and a previous study14 indicate that the reduction of either Exonucleases or Microprocessor levels, respectively, by RNAi knock-down causes a detectable reduction in the splicing of the miRNA-containing host introns. Co-transcriptional cleavage of introns may therefore facilitate splicing. This argues that the same intron transcript is subjected to both Drosha and splicing activities.
Note added in proof: J.M. Pawlicki and J.A. Steitz56 have recently presented complementary results suggesting that pri-miRNAs retained at transcription sites are processed to pre-miRNAs more efficiently than pri-miRNAs released into the nucleoplasm.
Methods
Primers
A list of oligonucleotides sequences is given in Supplementary Table 1 online.
Plasmid constructions
The miR-330 and let-7a3 sequences were PCR amplified using HeLa cell genomic DNA with the primer pairs 330β-F/330β-Rev and let7β-F/let7β-R, respectively. To generate the βInt2-miR-330 and βInt2-let-7a3 reporter constructs, PCR products were 5′ phosphorylated with polynucleotide kinase (Roche) in the presence of 1 mM ATP and blunt-end ligated to the βΔ5-8 vector backbone generated by long-range PCR with the Int2F/Int2a primer pair. We also carried out blunt-end ligation of the same miRNA fragments with different long-range PCR products resulting from PCR amplification of βΔ5-8 with the 3′B4/5′B10 primer pair to obtain the β3′-miR-330 and β3′-let-7a3 constructs. The mutant version of the miR-330-β construct was generated by blunt-end ligation of PCR product from amplification of the βInt2-miR-330 construct with 330m-fw/330m-rev oligonucleotides. To obtain wild-type and Δ hybrid-miR-223 constructs, a fragment (referred to as the miR-23a promoter) from −603 bp to +36 bp relative to the transcription-initiation site of pri-miR-23a~27a~24-2 was PCR amplified using miR-23-603fw/miR-23+36rev oligonucleotides and inserted into pcDNA 3.1 (+) plasmid (Invitrogen), lacking the T7 promoter and BGH poly(A) site, using BglII and NheI sites. DNA fragments from −92 (−50) and −42 (Δ) to +242 bp relative to the mature miR-223 sequence, obtained from genomic HeLa DNA by PCR amplification with mi-R223SL-50/ and miR223SL Δ/ miR-223 +200 oligonucleotides, respectively, were inserted between the NheI and EcoRI sites of pcDNA 3.1, containing the promoter region of pri-miR-23a~27a~24-2.
Cell culture and transfection procedure
HeLa cells were maintained in DMEM medium supplemented with 50 IU ml−1 of penicillin, 50 μg ml−1 streptomycin and 10% (v/v) FCS. Cells were cultured overnight at 37 °C in 5% CO2 to a confluency of 80–90% and then transiently transfected with test plasmid (10 μg) and Tat expression plasmid (1.5 μg) to allow activation of the HIV-1 promoter, using Lipofectamine 2000 (Invitrogen) reagent following the manufacturer’s instructions. RNA was extracted 24 h after transfection. RNAi procedures for XRN2, PMscl100, Drosha and DGCR8 knockdown have been described previously14,33,37,47.
The NB4 cell line was maintained in RPMI 1640 medium supplemented with 50 IU ml−1 of penicillin, 50 μg ml−1 streptomycin and 10% (v/v) FCS. Retinoic acid was purchased from Sigma and used at a concentration of 1 μM for the indicated times22.
Chromatin immunoprecipitation assays
We carried out ChIP assays as described previously48 with minor modifications. A total of 1 × 107 HeLa cells were fixed in 1% (v/v) formaldehyde for 10 min at room temperature (25–30 °C). Chromatin was sheared to an average size of 0.3–0.8 kb and immunoprecipitated with anti-Drosha (Abcam), anti–Pol II (N-20, Santa Cruz), anti-DGCR8 (Protein Tech Group), anti-PMscl100 (E. DijK, Radbound University) and rabbit polyclonal XRN2 antibody41.
We carried out ChIP analysis of transfected plasmids with the following modifications: HeLa cells were washed in PBS and resuspended in 4 ml of HLB/Nonidet P-40 buffer (0.14 M NaCl, 1.5 mM MgCl2, 10mM Tris-HCl, pH7.5, 0.5% (v/v) Nonidet P-40, 0.5 mM PMSF, 0.8μg ml−1 pepstatinA, 1μg ml−1 leupeptin). After incubating on ice for 5 min, nuclei were spun through 1 ml of underlayered HLB/Nonidet P-40 24% (w/v) sucrose buffer. Isolated nuclei were resuspended in 400 μl of nuclear lysis buffer (1% (w/v) SDS, 10mM EDTA and 50mM Tris, pH 8.1, 0.5 mM PMSF, 0.8μg ml−1 pepstatinA, 1μg ml−1 leupeptin) and incubated on ice for 10 min, and then the extracted chromatin was sonicated.
The immunoprecipitated DNA was recovered using the Qiaquick PCR purification kit (Qiagen). Quantitative PCR was performed with either a Rotorgene 3000 real-time PCR machine (Corbett Research) using Quantitec SYBR green (Qiagen) or in the presence of [α-32P]dATP as described previously49.
Northern blot analysis
Nuclear-cytoplasmic fractionation of HeLa RNA was carried out as previously described50. 30 μg of cytoplasmic HeLa and total NB4 (extracted by Trizol reagent) RNA was fractionated on 10% urea-polyacrylamide gel and transferred to a GeneScreen Plus membrane (Perkin Elmer). All the antisense oligonucletide probes (listed in Supplementary Table 1) were end-labeled using T4 polynucleotides Kinase (Bio Labs) following the manufacturer’s guidelines.
Biotinylated selection probes
EX2 and Bio-int2 riboprobe templates were generated by PCR using βInt2-miR-330 construct and either T7-EX2R/EX2F or T7-b2R/b2F oligo pairs respectively. Both riboprobe templates were then in vitro transcribed in the presence of 0.35 mM Biotin-16-UTP (Roche) as described previously51.
Hybrid selection circular rapid amplification of cDNA ends
HscRACE was performed as described33. EX2-H oligonucleotide was used to release the selected RNA from the streptavidin beads by RNase H (Roche) digestion. Reverse transcription of 5 μl ligated RNA mix was carried out using Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s guidelines in the presence of βEX2rev oligonucleotide. We PCR amplified 2 μl cDNA with βEX2rev/EX2fw divergent primers using Taq Polymerase (Invitrogen). Bands were excised from an agarose gel, cloned into the TOPO vector (Invitrogen) and then sequenced.
Reverse-transcription polymerase chain reaction
1 μg of RNA was reverse transcribed using Superscript III reverse transcriptase (Invitrogen) in a 20-μl reaction. Taq polymerase (Invitrogen) was used under standard conditions to amplify 2 μl of cDNA (20-25 cycles). For quantitative real-time PCR, 2 μl of cDNA was analyzed using a Rotorgene 3000 real-time PCR machine (Corbett Research) in the presence of Quantitec SYBR green (Qiagen).
Fractionation of chromatin-associated and nucleoplasmic transcripts
Nuclear fractionation and RNA extraction from both the nucleoplasmic and chromatin-associated pellet fractions were performed as previously described27,34.
Nuclear run on and hybrid selection nuclear run on
NRO and hybrid selection NRO analyses were carried out as described previously51.
Databases and EST analysis
The human miRNA list was retrieved from the miRBase registry release 10.1 (ref. 52). A BLAST search through the University of Calfornia in Santa Cruz (UCSC) web server53 and a miRGen query54 was initially carried out to identify the position of miRNA loci. After identifying the miRNA site, its position was compared with gene loci using RefSeq mRNA to identify miRNA matched with a protein-coding gene. For each sefseq entry, the UCSC database was used to indentify the features of all EST and splicing isoforms mapped to that region.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We are grateful to D. Cacchiarelli for the bioinformatics analysis and to M. Dye and S. West for helpful advice and discussion and for sharing unpublished data. M.M. was supported by an EMBO Long term fellowship. This work was partly supported by the 6th Framework Programme of the European Commission, SIROCCO and RIGHT Integrated Projects (LSHG-CT-2006-037900 and LSHB-CT-2004 005276) and Centro di Eccellenza BEMM to I.B. and by the ESF project “NuRNASu” to I.B. and N.J.P. The Proudfoot laboratory is supported by a Wellcome Trust Programme Grant.
Footnotes
Author Contributions
M.M. performed the experimental work except that M.B. carried out experiments on intergenic miRNA host genes together with F.P., and N.G. carried out experiments on exonuclease involvement. M.M., I.B., N.G. and N.J.P. wrote the paper. All authors read and agree with the paper’s contents.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 2.Basyuk E, Suavet F, Doglio A, Bordonné R, Bertrand E. Human let-7 stem-loop precursors harbour features of RNase III cleavage products. Nucleic Acids Res. 2003;31:6593–6597. doi: 10.1093/nar/gkg855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 5.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 6.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 7.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 8.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smalheiser NR. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 2003;4:403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu LH, et al. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Turris V, et al. TOP promoter elements control the relative ratio of intron-encoded snoRNA versus spliced mRNA biosynthesis. J Mol Biol. 2004;344:383–394. doi: 10.1016/j.jmb.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 19.Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 20.Richard P, Kiss AM, Darzacq X, Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol. 2006;26:2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang PK, et al. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 28.Gromak N, Talotti G, Proudfoot NJ, Pagani F. Modulating alternative splicing by cotranscriptional cleavage of nascent intronic RNA. RNA. 2007;14:359–366. doi: 10.1261/rna.615508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kluiver J, et al. Regulation of pri-microRNA BI transcription and processing in Burkitt lynphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 30.Thomson JM, et al. Extensive post-transcriptional regulation of microRNA and its implication for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West S, Gromak N, Proudfoot NJ. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 32.West S, Proudfoot NJ, Dye M. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H, Lin K. Excess of microRNAs in large and very 5′ biased introns. Biochem Biophys Res Commun. 2008;368:709–715. doi: 10.1016/j.bbrc.2008.01.117. [DOI] [PubMed] [Google Scholar]
- 36.Kampa D, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark TA, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 39.Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 41.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dye MJ, Gromak N, Haussecker D, West S, Proudfoot NJ. Turnover and function of noncoding RNA polymerase II transcripts. Cold Spring Harb Symp Quant Biol. 2006;71:275–284. doi: 10.1101/sqb.2006.71.040. [DOI] [PubMed] [Google Scholar]
- 43.Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 44.Kiss T. SnoRNP biogenesis meets Pre-mRNA splicing. Mol Cell. 2006;23:775–776. doi: 10.1016/j.molcel.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 45.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 46.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 47.Wagner EJ, Garcia-Blanco MA. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- 48.Forsberg EC, et al. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haussecker D, Proudfoot NJ. Dicer-dependent turnover of intergenic transcripts from the human β-globin gene cluster. Mol Cell Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- 56.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.