Abstract
Alternative splicing plays a major role in increasing proteome complexity and regulating gene expression. Here, we developed a new fluorescent protein-based approach to quantitatively analyze the alternative splicing of a target cassette exon (skipping or inclusion), which results in an open-reading frame shift. A fragment of a gene of interest is cloned between red and green fluorescent protein (RFP and GFP)-encoding sequences in such a way that translation of the normally spliced full-length transcript results in expression of both RFP and GFP. In contrast, alternative exon skipping results in the synthesis of RFP only. Green and red fluorescence intensities can be used to estimate the proportions of normal and alternative transcripts in each cell. The new method was successfully tested for human PIG3 (p53-inducible gene 3) cassette exon 4. Expected pattern of alternative splicing of PIG3 minigene was observed, including previously characterized effects of UV light irradiation and specific mutations. Interestingly, we observed a broad distribution of normal to alternative transcript ratio in individual cells with at least two distinct populations with ∼45% and >95% alternative transcript. We believe that this method is useful for fluorescence-based quantitative analysis of alternative splicing of target genes in a variety of biological models.
INTRODUCTION
Protein-coding eukaryotic genes possess exon–intron structure, and their primary transcripts undergo splicing—excision of introns from pre-mRNA to form mature mRNA consisting of exons only. Importantly, most genes of higher eukaryotes produce multiple isoforms of mature mRNA because of so-called alternative splicing, the process by which exons can be spliced in different arrangements (1). Alternative splicing provides a way to increase proteome complexity and to regulate gene expression in a tissue-, stage- or stimuli-dependent manner (2).
Regulation of alternative splicing is a complex process, which is still not completely understood. Short cis-acting sequence elements referred to as exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) or intronic splicing enhancers and silencers located within alternative exons and nearby introns, as well as adjacent constitutive exons, were found to play important roles in the alternative splicing regulation (1,3). Also, other structural features, such as RNA secondary structure and length of the exons, can be involved in determination of the alternative splicing patterns.
Classical methods to study alternative splicing include northern blot analysis and reverse transcriptase–polymerase chain reaction (RT–PCR). Modern high-throughput approaches, such as hybridization to oligonucleotide microarrays and large-scale sequencing of transcriptomes in combination with extensive bioinformatic examination, enable global analyses of alternative splicing (1,3).
Last decade, several approaches to analyze alternative splicing using green fluorescent protein (GFP) and other fluorescent proteins, e.g., red fluorescent protein (RFP), were developed (4–9). These methods enable visualization of results of alternative splicing in live cells and organisms at single-cell resolution. However, none of them gives the possibility of calculating relative amounts of mRNA species by measuring single cell fluorescence intensities (see ‘Results and Discussion’ section below).
Here, we suggest a new approach to analyze alternative splicing of a target cassette exon using two fluorescent proteins of different colors. A percentage of the full length and alternative (exon-skipping) forms can be estimated from the ratio of the fluorescent signals at single cell level.
MATERIALS AND METHODS
Genetic constructs
pSplPIG was made on the base of pTurboFP635-C vector (Evrogen) for expression of far-red fluorescent protein Katushka in mammalian cells. TagGFP-coding region was PCR amplified with primers 5′-GGATGTCGACGTGAATAGCGGGGGCGAGGAGCTG and 5′-GGATAGATCTCCTGTACAGCTCGTCCATGCC (restriction sites for SalI and BglII are underlined) and inserted in the corresponding sites (Sal1 and BamH1) of pTurboFP635-C vector. The obtained plasmid Katushka–TagGFP contained a polylinker between Katushka and TagGFP with sites for BglII and SalI.
The minigene of PIG3 was obtained by PCR amplification of exons 3–5 and the corresponding introns of PIG3 from HeLa genomic DNA using primers 5′-TCGCAGATCTCCGGAAATGTTCAGGCTGGAGACTAT and 5′-GGATGTCGACCGTCTGGGGCAGTTCCAGGACG. The PCR product (3.6 kb) was cloned into Katushka–TagGFP vector using BglII and SalI sites.
To prepare pCtrlPIG, a 592-bp cDNA fragment of PIG3 corresponding to exons 3–5 was amplified with the same specific primers as for pSplPIG (see above) on the template of HeLa cDNA. This fragment was cloned into Katushka–TagGFP vector using BglII and SalI sites.
Mutagenesis
Our mutagenetic efforts were based on the previously published work (10), where 12 bp segments of exon 4 were systematically replaced with a heterologous sequence TCAGCATGACTC lacking splicing enhancer or silencer activities. However, this sequence contains stop codon (underlined), which is unacceptable for our method, and also can result in artifacts due to nonsense-mediated decay of mRNA with a premature stop codon. Thus, to replace target regions in PIG3 exon 4, we used sequence TCAGCATGTCTC with no stop codon.
In mutants MutE and MutH, the following regions of exon 4 were substituted for TCAGCATGTCTC: AAGAACGTCAAC (positions 49–60 of exon 4, MutE) or GTTCTCTATGGT (positions 87–98 of exon 4, MutH). In mutant MutCU, we introduced multiple silent substitutions (underlined) into regions 86–133 of exon 4:
GTATTA TAT GGCTTA ATG GGT GGC GGA GAT ATT AAC GGA CCATTG TTC.
To clone the mutated sequences into pSplPIG, we used BamHI and ApaI restriction sites, which are in proximity to the target region of the PIG3 gene (upstream and downstream, respectively).
Cell culture
Human embryonic kidney HEK293T cell line was cultured in DMEM (PanEco) with 10% FBS (Sigma) at 37°C in 5% CO2 atmosphere. Transient transfections were performed with FuGene 6 reagent (Roche) according to the manufacturer's protocol. Cells were analyzed by fluorescence microscopy and flow cytometry 24–48 h after transfection.
For UV treatment, cells before transfection were placed in PBS and exposed to Philips G8T5 germicidal lamp (254 nm) for 90 s at about 1 m distance (dose of about 100 J/m2).
Fluorescence microscopy
Fluorescence microscopy of live cells was performed using Leica AF6000 LX imaging system equipped with a Photometrics CoolSNAP HQ CCD camera and a 120 W HXP short arc lamp (Osram). Green and red fluorescence images were acquired using ×10 objective and standard filter sets: GFP (excitation BP470/40, emission BP525/50) and TX2 (excitation BP560/40, emission BP645/75).
Flow cytometry
Flow cytometry of live cells was performed using Beckman Coulter Cytomics FC500 equipped with 488 nm laser. Fluorescence was detected in green (FL1, 510–540 nm) and red (FL4, 660–690 nm) channels. HEK293T cells expressing either Katushka or TagGFP (Evrogen) were used as controls to check for cross talk of TagGFP signal into red channel and vice versa. Digital data for calculations of alternative transcript percentage were extracted using Weasel software and processed by Origin software.
RESULTS AND DISCUSSION
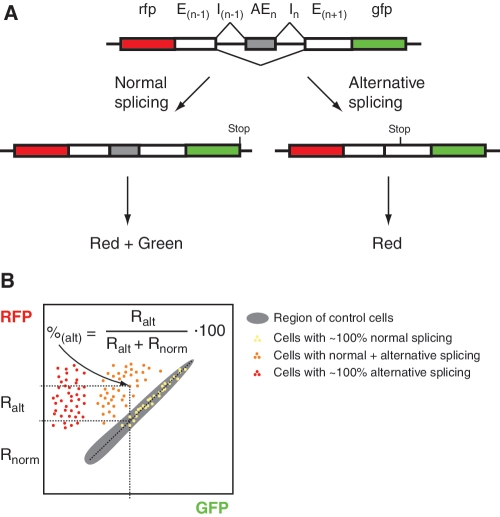
The proposed method is aimed to analyze alternative splicing of cassette exons, skipping or including of which results in an open reading frame shift (i.e. exon length is not a multiple of 3). Two fluorescent proteins of clearly different colors are used for quantitative analysis at single live cell level. A fragment of target gene (minigene) consisting of an alternative exon (AEn) of interest flanked by adjacent upstream and downstream introns (I) and constitutive exons (E) is cloned between RFP- and GFP-encoding sequences in the following order: rfp-E(n−1)-I(n−1)-AEn-In-E(n+1)-gfp (Figure 1A) so that the ORF is conserved from RFP to GFP. Thus, translation of the normally spliced full-length transcript results in a RFP-tORF-GFP fusion protein. In contrast, alternative exon skipping results in a frame shift within exon E(n+1) and thus leads to a truncated protein RFP-tORF without GFP.
Figure 1.
Outline of the proposed method. (A) Schematic representation of the reporter, which includes target gene fragment (an alternative exon with adjacent introns and constitutive exons) flanked by RFP- and GFP-coding regions. Alternative splicing results in two mRNA species. Translation of the normal full-length transcript (left) produces RFP and GFP, whereas translation of the alternative short transcript (right) results in RFP only. (B) Scheme of expected flow cytometry results (GFP-RFP bivariate plot). Gray area shows control cells expressing normal transcript without introns. Cells with only normal splicing, only alternative splicing and both types of splicing of the target gene are shown as yellow, red and orange dots, respectively. A percentage of normal and alternative transcripts in each cell can be estimated using designated equation.
Cells expressing this construct develop different levels of red and green fluorescence (which can be quantified using fluorescence microscopy or flow cytometry), and the ratio of red to green can be used to estimate a proportion of normal and alternative transcripts. To this end, a control construct encoding RFP-tORF-GFP without introns should be used. Expression of this control provides a reference ratio between the red and the green signals observed in a particular model and detection settings when only normal transcript is present. In the experimental construct undergoing alternative splicing, additional red signal, which is above the estimated control red/green ratio, corresponds to the alternative transcript.
Experimental results can be represented as a plot where each cell gives a dot of particular intensities in green (abscise axe) and red (ordinate axe) channels (Figure 1B). On this plot, the control cells would form a diagonal cluster, whereas experimental cells would lie either on or above this diagonal. Portions of normal and alternative transcripts in each cell can be estimated using a simple calculation using its position relative to the control diagonal (Figure 1B).
To test this new method, we analyzed alternative splicing of human PIG3 (p53-inducible gene 3) gene. This gene represents an interesting example of a regulatable alternative splicing. Expression of PIG3 produces two splice variants: major full-length transcript consisting of all 5 exons and minor short transcript lacking exon 4 of 197-bp length (10). Absence of exon 4 induces a frame shift in exon 5.
In this work, we used far-red fluorescent proteins Katushka and GFP TagGFP. These fluorescent proteins provide bright and spectrally non-overlapping signals. In the previous work, we showed that the coding region of Katushka contains a strong donor splice site, which can interfere with target gene splicing (11). Therefore, we used a mutated Katushka gene with altered codon usage that does not possess splice sites and, thus, can be used to analyze splicing events (11). We cloned PIG3 minigene (exons 3–5 and corresponding introns) between Katushka and TagGFP-coding sequences (plasmid pSplPIG) in accordance to the scheme depicted in Figure 1A. Also, a control plasmid pCtrlPIG corresponding to the normally spliced transcript (Katushka-exons3–5-TagGFP) was constructed. HEK293T cells were transfected with these plasmids and analyzed using fluorescence microscopy and flow cytometry. Cells expressing pCtrlPIG demonstrated bright green and red fluorescence; the ratio between red and green signals did not vary much from cell to cell. Therefore, overlay of the images in green and red channels gave mainly yellow cells (Figure 2A), and flow cytometry showed that these cells form a quite narrow diagonal on the red–green bivariate plot (Figure 3A).
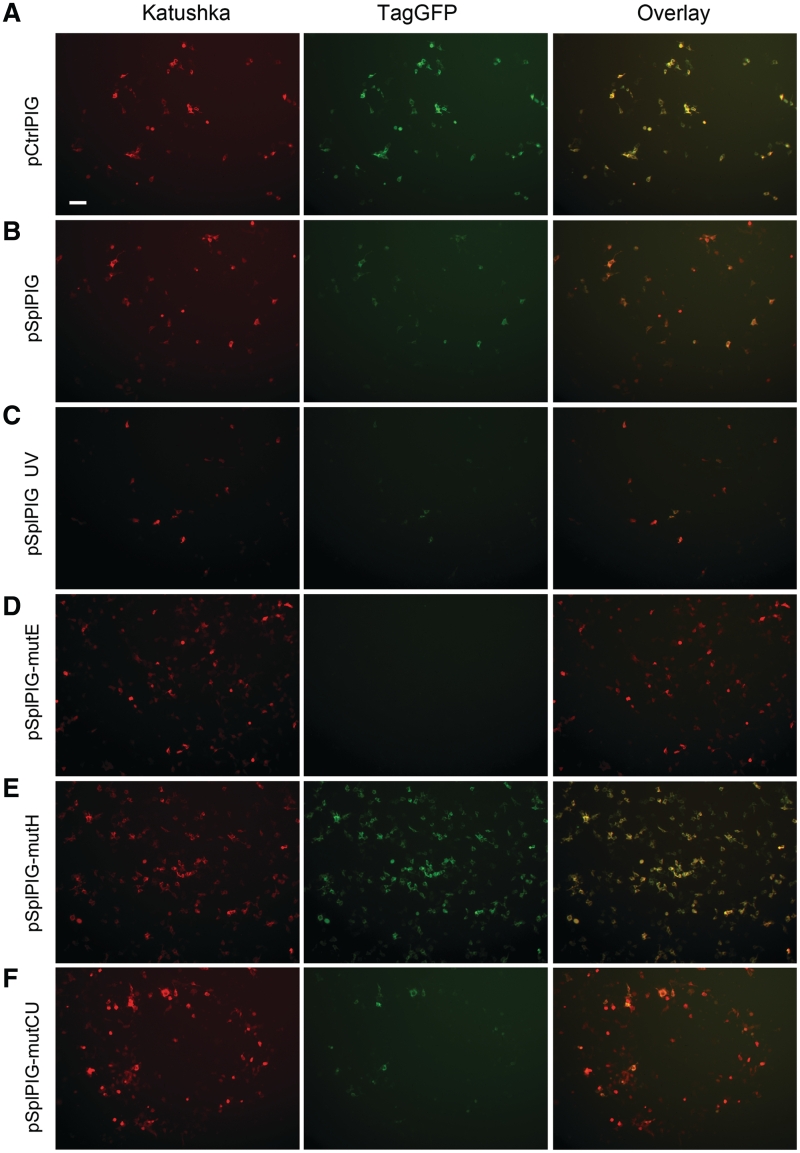
Figure 2.
Fluorescence microscopy of HEK293T cells transiently expressing target vectors. Left column, red channel; middle, green channel and right, overlay. All cells were imaged using identical settings. Scale bar 100 µm. (A) pCtrlPIG; (B) pSplPIG; (C) pSplPIG, cells were treated with UV; (D) pSplPIG-mutE; (E) pSplPIG-mutH; and (F) pSplPIG-mutCU.
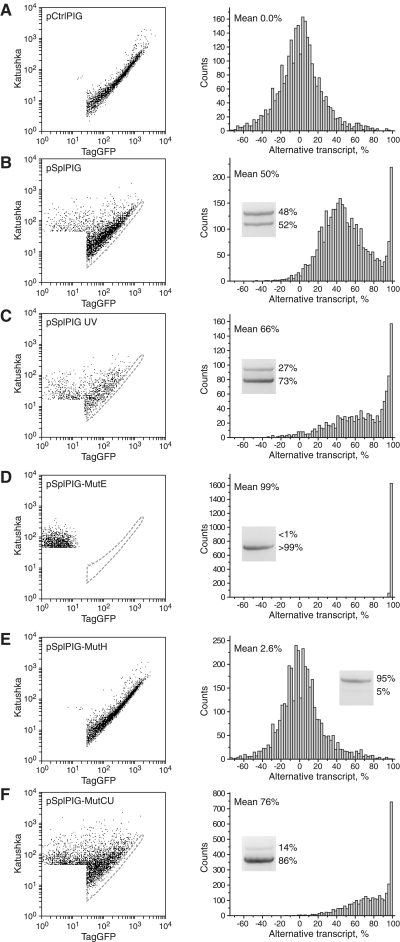
Figure 3.
Flow cytometry analysis of HEK293T cells transiently expressing target vectors. Left panels: dot plots of cell fluorescence in green (TagGFP) and red (Katushka) channels. All samples were analyzed using the same settings of the flow cytometer. Area of non-transfected and low fluorescent cells is not shown and not taken for analysis. Area corresponding to the pCtrlPIG-expressing cells is outlined by dashed gray line on each plot. Right panels: quantitative analysis of the corresponding left plots. For each cell, a percentage of alternative transcript was estimated, and these data were used to build a histogram of cell distribution in the sample. Cells with seemingly negative percentage of alternative transcript come from the area below the middle line of the control sample. Insets show results of RT–PCR analysis of the corresponding cDNA samples. Upper band (∼1100 bp) corresponds to the normal PIG3 transcript, lower band (∼900 bp) corresponds to the alternative transcript without exon 4. The percentage of each band was calculated by densitometry with correction for the difference in size of the DNA fragments. (A) pCtrlPIG; (B) pSplPIG; (C) pSplPIG, cells were treated with UV; (D) pSplPIG-mutE; (E) pSplPIG-mutH; and (F) pSplPIG-mutCU.
In contrast, transfection with pSplPIG revealed unexpected cell-to-cell variation in a red/green ratio. Side-by-side analysis of pSplPIG- and pCtrlPIG-expressing cells under the same detection settings showed that some pSplPIG cells were similar to the control cells, some others were almost purely red, and majority of pSplPIG cells gave various relative intensities of red and green signals (with a clear prevalence of red over green fluorescence compared with pCtrlPIG cells) (Figure 2B). We concluded that levels of PIG3 minigene alternative splicing in individual HEK293T cells can vary significantly—from nearly 100% normal transcript (‘yellow’ cells) to nearly 100% alternative transcript (‘red’ cells), but most cells produce both transcripts in various proportions (‘orange’ cells).
Quantitative analysis of flow cytometry data confirmed a broad distribution of the proportion between normal and alternative transcripts in pSplPIG cells (Figure 3B). Two clear peaks at ∼45% and >95% of alternative transcript can be recognized. Thus, at least two distinct populations of cells with different levels of alternative splicing of the target gene exist in HEK293T. A mean percentage of pSplPIG alternative form was estimated to be 50%. To check this value by an independent method, we isolated RNA from HEK293T transfected with pSplPIG, generated cDNA, and performed PCR with primers corresponding to Katushka and TagGFP sequences. As expected, this amplification resulted in two bands corresponding to the normally and alternatively spliced transcripts (Figure 3). Densitometry analysis showed lower band (alternative transcript) to constitute 52% of the total, which is in a good agreement with flow cytometry data.
The production of the PIG3 alternative transcript was found to be enhanced by irradiation of cells with UV light (10). Thus, we tested the effect of UV illumination on pSplPIG splicing. In line with literature data (10), a considerable increase in the percentage of the alternative transcript was observed (Figures 2C and 3C).
Earlier mutagenesis studies on PIG3 minigene revealed regions within exon 4 acting as strong ESE or ESS (10). We generated two mutants, pSplPIG-mutE and pSplPIG-mutH, corresponding (with some modifications, see Materials and Methods for details) to two extreme mutants dubbed e and h in the original publication (10), which produce nearly exclusively alternative and normal transcripts, respectively. pSplPIG-mutE-expressing cells developed only red fluorescence, which corresponds to the presence of alternative and absence of the normal transcripts (Figures 2D and 3D). In contrast, transfection with pSplPIG-mutH resulted in ‘yellow’ cells, which practically overlap with the control pCtrlPIG-expressing cells (purely normal splicing) on the bivariate plot (Figures 2E and 3E). Thus, both mutants showed expected behavior.
We also generated a mutant pSplPIG-mutCU with altered codon usage, which contains no amino acid substitutions but many silent mutations within exon 4 nucleotides 86–133 [designated h–k in (10)]. This region was shown to be ESS (10), and we thus aimed to generate PIG3 variant with no exon 4 skipping. Unexpectedly, pSplPIG-mutCU demonstrated a considerable enhancement of exon 4 skipping (Figures 2F and 3F). Interestingly, the splicing pattern of this mutant was very similar to that observed for UV-treated pSplPIG cells. We believe that pSplPIG-mutCU can potentially be used as a model of UV-induced changes of PIG3 alternative splicing (but without UV irradiation) to study biochemical consequences of this phenomenon.
In conclusion, our method provided an adequate evaluation of PIG3 minigene splicing including expected changes in response to UV and introduced mutations. Moreover, analysis at single-cell level made it possible to reveal a strong heterogeneity of cells with respect to PIG3 alternative splicing. The reasons for this heterogeneity are unclear and call for further investigation. Generally, heterogeneity within seemingly uniform cell populations recently became the focus of extensive research (12–14). It can be revealed by cell staining for several independent markers (15,16). Heterogeneity of alternative splicing can add a new dimension to these studies.
We believe that the new method is potentially useful for the analysis of alternative splicing of target genes in a variety of biological models—cell lines and whole organisms—where fluorescence microscopy and/or flow cytometry can be used to detect fluorescence. The method possesses common drawbacks of GFP-based techniques such as the necessity to (over)express an exogenous gene (that can affect cell physiology including splicing), and potentially altered stability of chimeric mRNA with and without alternative exon. However, worldwide practice of applications of fluorescent proteins shows that it can be used safely in most biological models. Advantages of the method proposed can be summarized as follows:
A new scheme of arrangement of two fluorescent proteins is applied. The main novelty of the scheme is that RFP is encoded in both normal and alternative transcripts. This design allows quantitative analysis of alternative splicing in individual cells by decomposing red signal into two portions corresponding to alternative (RFP) and normal (RFP+GFP) transcripts using a simple control sample (RFP+GFP). In contrast, previously published methods do not allow calculating the percentage of transcripts from fluorescence intensities even when two fluorescent proteins are used (5,6,8,9). A quantitative analysis of a cassette exon skipping within single cells using EGFP and DsRed was claimed by Orengo et al. (7). However, this method is based on the comparison of fluorescence intensity of a cell in green channel with that of the same cell in red channel as assessed by fluorescence microscopy or flow cytometry. In fact, such direct comparison of the green fluorescence signal to the red one to estimate molar ratio between EGFP and DsRed (or any other fluorophores) seems to be incorrect for many reasons including the following: (a) different fluorophores possess different intrinsic (per molecule) brightness, (b) excitation filters (lasers) and emission filters fit the excitation–emission spectra of the fluorophores differently, (c) intensity of excitation light (e.g. from a mercury lamp) can differ drastically in different spectral regions and (d) the detector can possess different sensitivity in different spectral regions.
A large fragment of the unaltered target gene including an alternative exon with flanked introns and constitutive exons is taken for analysis. This ensures inclusion of nearly all native regulatory elements, which are usually concentrated not only within alternative exons and flanked introns but also can be found in adjacent constitutive exons (3). At the same time, expression cassettes in some existing methods of fluorescent protein-based analysis of alternative splicing do not include adjacent constitutive exon(s) or contain fluorescent protein-coding inserts into alternative exons (5,9). In some cases, this can affect the regulation of alternative splicing.
In addition to cassette exons, our method can be potentially adapted for other alternative splicing types such as alternative 5′- or 3′- splice sites, intron retention and mutually exclusive exons. The only requirement is a frame shift in the alternative transcript relative to the normal one. The frame shift was found to occur in more than one-third of all alternative splicing events in human and mouse (1,17). Thus, the method suggested is applicable for a large number of genes. For other genes, artificial alteration of the size of alternative regions by 1–2 nucleotides can be used to induce the required frame shift (6,7).
FUNDING
Molecular and Cell Biology program of the Russian Academy of Sciences; Russian Foundation for Basic Research (10-04-01042-a and 11-04-12187-ofi-m); and Ministry of Education and Science of the Russian Federation (16.740.11.0367 and 16.512.11.2139). Funding for open access charge: Molecular and Cell Biology program of the Russian Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell. Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 3.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat. Methods. 2006;3:909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- 6.Newman EA, Muh SJ, Hovhannisyan RH, Warzecha CC, Jones RB, McKeehan WL, Carstens RP. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orengo JP, Bundman D, Cooper TA. A bichromatic fluorescent reporter for cell based screens of alternative splicing. Nucleic Acids Res. 2006;34:e148. doi: 10.1093/nar/gkl967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno G, Hagiwara M, Kuroyanagi H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 2008;22:360–374. doi: 10.1101/gad.1620608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl Acad. Sci. USA. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholls CD, Beattie TL. Multiple factors influence the normal and UV-inducible alternative splicing of PIG3. Biochim. Biophys. Acta. 2008;1779:838–849. doi: 10.1016/j.bbagrm.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Gurskaya NG, Staroverov DB, Fradkov AF, Lukyanov KA. Coding region of far-red fluorescent protein Katushka contains a strong donor splice site. Russ. J. Bioorg. Chem. 2011;37:380–382. doi: 10.1134/s1068162011030071. [DOI] [PubMed] [Google Scholar]
- 12.Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 13.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slack MD, Martinez ED, Wu LF, Altschuler SJ. Characterizing heterogeneous cellular responses to perturbations. Proc. Natl Acad. Sci. USA. 2008;105:19306–19311. doi: 10.1073/pnas.0807038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh DK, Ku CJ, Wichaidit C, Steinenger RJI, Wu LF, Altschuler SJ. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol. Syst. Biol. 2010;6:369. doi: 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]