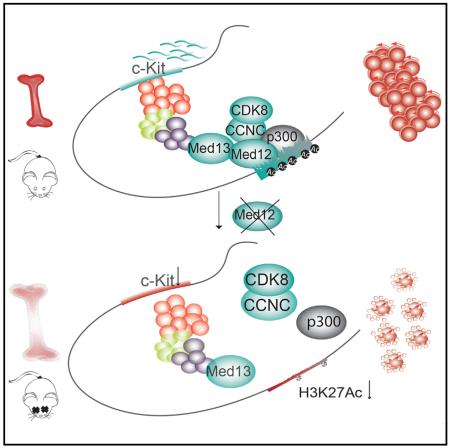
SUMMARY
Hematopoietic-specific transcription factors require coactivators to communicate with the general transcription machinery and establish transcriptional programs that maintain hematopoietic stem cell (HSC) self-renewal, promote differentiation, and prevent malignant transformation. Mediator is a large coactivator complex that bridges enhancer-localized transcription factors with promoters, but little is known about Mediator function in adult stem cell self-renewal and differentiation. We show that MED12, a member of the Mediator kinase module, is an essential regulator of HSC homeostasis, as in vivo deletion of Med12 causes rapid bone marrow aplasia leading to acute lethality. Deleting other members of the Mediator kinase module does not affect HSC function, suggesting kinase-independent roles of MED12. MED12 deletion destabilizes P300 binding at lineage-specific enhancers, resulting in H3K27Ac depletion, enhancer de-activation, and consequent loss of HSC stemness signatures. As MED12 mutations have been described recently in blood malignancies, alterations in MED12-dependent enhancer regulation may control both physiological and malignant hematopoiesis.
Graphical Abstract
INTRODUCTION
Stem cells rely on the finely coordinated activity of cell-specific transcription factors to self-renew and differentiate (De Los Angeles et al., 2015; Spitz and Furlong, 2012). Key transcription factors, such as GATA2, RUNX1, GFI1, or TAL1, are required to preserve hematopoietic stem cell (HSC) function (Orkin and Zon, 2008; Wilson et al., 2010). HSC function requires a tightly regulated network of transcription factors to guarantee homeostasis and prevent transformation (Lara-Astiaso et al., 2014; Rossi et al., 2012). Transcription factors use coactivators to communicate with the general transcription machinery and ensure that biological inputs received from signaling cascades are translated into specific gene expression programs (Spiegelman and Heinrich, 2004; Weake and Workman, 2010). The Mediator complex is a pivotal coactivator of transcription factor activity that acts as a molecular bridge between transcription factors at enhancers and RNA polymerase II at promoters (Allen and Taatjes, 2015). Mediator is a large macromolecular complex arranged in four modules, the head, the middle, the tail, and the kinase module, the latter being comprised by MED12, MED13, CDK8, and CYCLIN C (Tsai et al., 2014).
An essential feature of Mediator is the ability to regulate its function by modulating its subunit composition (Allen and Taatjes, 2015). However, the contribution of each subunit to the function of the complex remains largely unexplored. Mediator plays a role in various fundamental processes, such as transcription initiation (Wang et al., 2005), pause release (Galli et al., 2015), elongation (Takahashi et al., 2011), and chromatin architecture (Kagey et al., 2010; Ørom et al., 2010). The kinase module reversibly interacts with the Mediator core through MED13, causing a structural shift and further modulation of its function (Davis et al., 2013; Knuesel et al., 2009a). While most subunits of Mediator are evolutionarily conserved (Malik and Roeder, 2010), three members of the kinase module evolved to have paralogs in vertebrates (MED12L, MED13L, and CDK19) (Bourbon, 2008). The kinase module functions in a context-dependent manner as an activator or repressor of Mediator function (Galbraith et al., 2013; Pelish et al., 2015). While most functions of the kinase module are attributed to CDK8/CDK19 kinase activity (Knuesel et al., 2009b), additional roles of this module remain unknown. Whereas each subunit of the complex can interact with different transcription factors (Borggrefe and Yue, 2011), transcription factor interactions in the kinase module have been largely attributed to MED12 (Malik and Roeder, 2010). However, little is known about the role of MED12 (or any other Mediator subunit) in adult stem cell and specifically HSC function. The first indication of a function for MED12 in hematopoiesis came from zebrafish studies, showing a potential role in myelopoiesis (Keightley et al., 2011). The recent identification of MED12 mutations in cancer, including leukemia (KÄmpjÄrvi et al., 2016), further supports this notion.
We investigated the role of MED12 in hematopoietic stem and progenitor cells (HSPCs) in vivo using several hematopoieticspecific Med12 knockout animal models. We show that MED12 is essential for HSC homeostasis as Med12 deletion led to HSPC loss, bone marrow failure, and rapid lethality. MED12-deficient HSPCs promptly lost key HSC gene expression signatures and were highly apoptotic. These effects appear independent from MED12 function within the Mediator kinase module, as MED13, CYCLIN C, and CDK8 were dispensable for normal hematopoiesis. Finally, we characterize a mechanistic function for MED12 in cooperation with P300 in the maintenance of the active state of hematopoietic enhancers. Altogether, we demonstrate an indispensable role for MED12 in vivo in adult stem cell homeostasis, and we describe MED12-regulated mechanisms in hematopoiesis.
RESULTS
MED12 Is Essential for Hematopoiesis
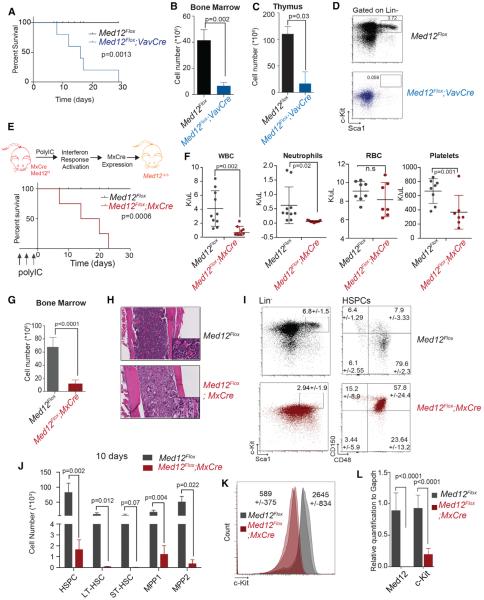
To interrogate the function of MED12 in the hematopoietic system, we initially used Med12Flox mice (Figure S1A) (Rocha et al., 2010) and conditionally deleted Med12 with Vav1-Cre recombinase, which is active as early as embryonic day (E)10.5 (Stadtfeld and Graf, 2005). Med12Flox;VavCre animals were born at reduced numbers as indicated by the perturbation of Mendelian ratios at birth (Figure S1B). Med12Flox;VavCre animals died within 2 weeks of birth (Figure 1A) and showed a severe reduction of bone marrow and thymus cellularity (Figures 1B and 1C), suggesting aberrant hematopoiesis. Flow cytometric analysis of bone marrow revealed that HSPCs were dramatically affected (Figure 1D). A significant decrease in HSPC numbers was readily observed on fetal livers at E15.5 (Figures S1C and S1D), suggesting that MED12 also is required for adequate fetal hematopoiesis.
Figure 1. MED12 Deletion Leads to Bone Marrow Failure and Rapid Animal Lethality.
(A) Kaplan-Meier curve plotting survival of Med12Flox;Vav-Cre and control mice (n = 8) is shown.
(B and C) Total cell counts in bone marrow (B) and thymus (C) of Med12Flox;Vav-Cre and control animals (n = 3) 1 week after birth are shown.
(D) Representative immunophenotypic analysis of bone marrow of the indicated mice (n = 3) is shown.
(E) Kaplan-Meier curve plotting survival of Med12Flox;Mx-Cre and Med12Flox animals (n = 4) is shown.
(F) Peripheral blood counts of Med12Flox;Mx-Cre and controls 10 days after first pI:pC injection (n = 8–10) are shown.
(G) Bone marrow counts (femura, tibia, and hip) of Med12Flox;Mx-Cre animals and Med12Flox 10 days after pI:pC injection are shown.
(H) H&E staining of bone sections is shown.
(I) Immunophenotypic of HSPCs of indicated mice 4 days after second pI:pC injection (n = 4) is shown.
(J) Absolute numbers of HSC populations are shown (HSPCs: Lin−, Sca-1+, and c-Kit+; LT-HSC: CD150+ and CD48−; ST-HSC: CD150− and CD48−; MPP1: CD150+ and CD48+; and MPP2: CD150− and CD48+).
(K) Mean geometric frequency of c-KIT surface marker 4 days after second pI:pC injection (n = 4) (p < 0.0005) is shown.
(L) Expression of Med12 and c-Kit in HSPCs 4 days after two pI:pC injections (n = 4).
Error bars represent mean ± SD; p values were determined with two-tailed Student's t tests. See also Figures S1 and S2.
Expression analysis showed that Med12 is expressed in all hematopoietic populations and presents the highest protein levels in mouse HSPCs (Figures S1E and S1F). To test whether MED12 is required for adult hematopoiesis, we crossed Med12Flox animals to the Mx1Cre strain (Figure 1E), where Cre-recombinase expression is induced by injection of polyinosine-polycytidine (pI:pC) through interferon-α (IFN-α) response (Kühn et al., 1995). Recombination of the Med12 floxed allele occurred within 4 days post-injection (Figure S1G), and MED12 protein was undetectable in the thymus from MED12-deficient animals (Figure S1H). Adult mice lacking Med12 expression showed low white blood cell and platelet counts and died within 3 weeks post-deletion (Figure 1F). Bone marrow cellularity was rapidly reduced (Figure 1G) and, similarly, the size of thymus and spleen (both dependent on progenitor output from the bone marrow) (Figures S1I and S1J). Histological analysis confirmed bone marrow hypocellularity (Figure 1H) and aberrant structures in primary hematopoietic organs (Figure S1K). Med12 deletion almost completely obliterated HSPCs within 4 days post pI:pC, and, within 10 days, HSPCs were absent (Figures 1I and S1M).
We used SLAM markers (CD150 and CD48) to subdivide the HSPC subset into long-term HSC (LT-HSC), short-term HSC (ST-HSC), and multi-potent progenitor (MPP1 and MPP2) populations. Immunophenotypic analysis showed that all populations were significantly depleted already 4 days after pI:pC (Figures 1J and S1L). Moreover, MED12-deficent mice showed a decrease of c-KIT both on the cell surface (Figure 1K) and at the mRNA level (Figure 1L). c-KIT is the surface receptor of stem cell factor (SCF), a cytokine essential for HSC self-renewal, growth, and survival (Domen and Weissman, 2000). These data show that MED12 loss in the hematopoietic system leads to severe bone marrow failure and consequent lethality.
MED12 Function in Hematopoiesis Is Cell Autonomous
To assay cell-intrinsic effects of Med12 deletion in HSC function, we performed bone marrow transplantations with bone marrow cells from Med12Flox;MxCre and littermate controls 1 week after pI:pC injection. Lethally irradiated recipients receiving MED12-deficient bone marrow cells died within 2 weeks, suggesting a failure in engraftment and bone marrow repopulation in the absence of MED12 (Figure S2A). Competitive bone marrow transplantations were performed by mixing equal numbers of bone marrow cells from CD45.2+ Med12Flox;MxCre mice or CD45.2+ Med12Flox littermates and CD45.1+ wild-type bone marrow before pI:pC injection (Figure S2B) and transplanting them into lethally irradiated recipient animals. Med12 deletion was induced upon confirmation of comparable engraftment rates, and chimerism was monitored by immune-phenotypic analysis of CD45.1+ and CD45.2+ over 20 weeks. As shown in Figure S2C, MED12-deficient (CD45.2+) cells were rapidly out-competed by co-transplanted CD45.1+ wild-type cells. Myeloid cells were the first to disappear from peripheral blood, as they have a shorter lifespan, and, over the course of 20 weeks, lymphoid populations also were reduced albeit with slower kinetics (Figure S2C), indicating that no de novo hematopoiesis originated form MED12-deficient bone marrow cells.
We analyzed the proliferation and differentiation potential of Med12Flox cells in colony formation assays. Bone marrow cells 10 days after Med12 deletion were unable to form colonies compared to control littermates (Figure S2D). To rule out cell-extrinsic effects potentially arising from in vivo Med12 deletion, we treated HSPCs from Med12Flox;MxCre mice with IFN-α ex vivo at the time of plating in colony-forming unit (CFU) assays, and we confirmed that MED12-deficient cells had a reduced ability to form colonies (Figure S2E). Similarly, HSPCs failed to grow in ex vivo liquid culture cell upon MED12 deletion (Figure S2F). These results demonstrated that defects caused by MED12 loss occur cell autonomously.
MED12 is highly conserved between species with mouse and human MED12 protein showing a 97% homology (protein blast, e value = 0.0). We wondered whether MED12 loss would have the same consequences in human HSPC as seen in mice. We used gene-specific small hairpin RNAs (shRNAs) to knock down MED12 in mobilized CD34+ peripheral blood stem cells, and we performed specific colony formation assays that demonstrated that MED12 knockdown led to a significant reduction of the colony-forming ability of human HSPCs (Figure S2G), suggesting that MED12 loss of function in hematopoiesis is conserved among vertebrates.
MED12 Loss Does Not Universally Affect Cell Survival
To investigate if MED12 loss of function universally affects cell viability and growth, we immortalized mouse embryonic fibroblasts (MEFs) from Med12Flox animals and deleted Med12 in vitro. Monitoring cell growth showed that MED12 deletion did not affect MEF growth rate (Figure S2H), which could grow indefinitely in culture (data not shown). Mouse embryonic stem cells (mESCs) from germline MED12-deficient mice also were able to grow in vitro, form defined colonies (Figure S2I), continue to express key ESC pluripotent markers (Figure S2I) when maintained on feeders under pluripotency conditions, and contribute to the three embryonic germ layers in tetraploid complementation studies (Rocha et al., 2010).
Finally, we crossed Med12Flox mice to a tamoxifen-inducible Cre-recombinase estrogen receptor-T2 (CreER) animal strain (Ventura et al., 2007). CreER-Med12-deleted mice also showed a striking reduction of bone marrow HPSCs (data not shown). Immunofluorescence experiments of skin sections revealed that hair follicle stem cells did not apoptose in response to MED12 deletion, while epidermal progenitor cells showed reduced expression of the proliferation marker Ki-67 (Figure S2J). Histological analysis of a number of additional tissues did not reveal dramatic effects at a macroscopical scale (data not shown), suggesting that MED12 is not universally required for cell survival.
Kinase Module-Independent Activity of MED12 in the Hematopoietic System
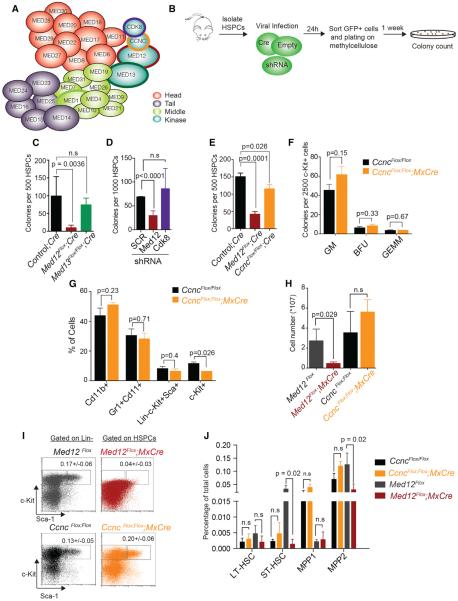
Previous reports had shown that MED1, a member of the Mediator core, is dispensable for HSC maintenance (Stumpf et al., 2010). As MED12 is part of the kinase module of Mediator (Figure 2A), we tested whether deletion of other members of this module would result in similar phenotypes. First we deleted MED13, which anchors the kinase module to the core Mediator complex (Knuesel et al., 2009a). We transduced HSPCs from Med13Flox/Flox mice (Grueter et al., 2012) with Cre-IRES-GFP recombinase retrovirus, sorted infected cells, and performed CFU assays (Figure 2B). Surprisingly, and unlike MED12 loss, MED13 depletion did not significantly affect colony formation ability of mouse HSPCs in these conditions (Figures 2C and S3A–S3C). Med13 deletion was confirmed using PCR on genomic DNA and qRT-PCR (Figures S3D and S3E).
Figure 2. Kinase Module-Independent Activity of Med12 in the Hematopoietic System.
(A) Schematic of the Mediator complex is shown.
(B) Methylcellulose CFU assay strategy is shown.
(C) CFU assay with HSPCs from control, Med13Flox/Flox, and Med12Flox mice infected with Cre virus is shown.
(D) CFU assay in wild-type mouse HSPCs infected with indicated shRNAs is shown.
(E) CFU assay using HSPCs from controls, CcncFlox/Flox, and Med12Flox mice transduced with Cre virus is shown.
(F) CFU assay with c-Kit+-enriched cells from CcncFlox/Flox and CcncFlox/Flox;MxCre mice 10 days after pI:pC injection is shown.
(G) FACS surface marker analysis of colonies from (F) is shown.
(H) Total bone marrow cellularity of indicated mice is shown.
(I) Immunophenotypic analysis of control, CncFlox/Flox;MxCre (10 days after pI:pC), and Med12Flox;MxCre (4 days after pI:pC) is shown.
(J) Frequencies of HSPC populations in indicated mice.
Data represent mean ± SD (n = 3; p values were determined with two-tailed Student's t tests). See also Figure S3.
Since most regulatory functions of the kinase module have been attributed to its kinase activity (Allen and Taatjes, 2015), we wondered whether the MED12-deletion phenotype could be caused by the loss of CDK8 activity. To test this, we isolated HSPCs and infected them with lentiviruses expressing shRNAs against Cdk8 or Med12. CFU assays revealed that a reduction of Cdk8 expression did not affect HSPC colony formation ability (Figures 2D, S3F, and S3G).
MED12 activates CDK8 through binding CYCLIN C (Turunen et al., 2014), the only kinase module component without a paralog. Therefore, deletion of CYCLIN C abrogates binding between MED12 and CDK8 (Li et al., 2014). To rule out that the lack of phenotype upon Cdk8 knockdown was due to residual kinase activity, we purified HSPCs from CcncFlox/Flox mice and deleted CYCLIN C with Cre recombinase as described above (Figure S3H). Similar to Cdk8 silencing and Med13 deletion, Ccnc deletion had no significant effects on the ability of HSPC cells to form colonies (Figure 2E). We confirmed these findings in CFU assays with c-KIT-positive enriched cells isolated from CcncFlox/Flox;MxCre after pI:pC injection (Figures 2F and 2G). Analysis of bone marrow cells from CcncFlox/Flox; MxCre after pI:pC also showed that, contrary to MED12 deletion, loss of CYCLIN C in vivo did not lead to bone marrow failure (Figures 2H and S3I) and had no significant effects on the HSPC population (Figures 2J and S3J) or c-KIT levels (Figures 2I and S3K). This detailed analysis of the Mediator kinase module suggested that MED12 could have additional functions independent of its role within the kinase module.
MED12 Loss Does Not Disrupt Mediator Core Structure
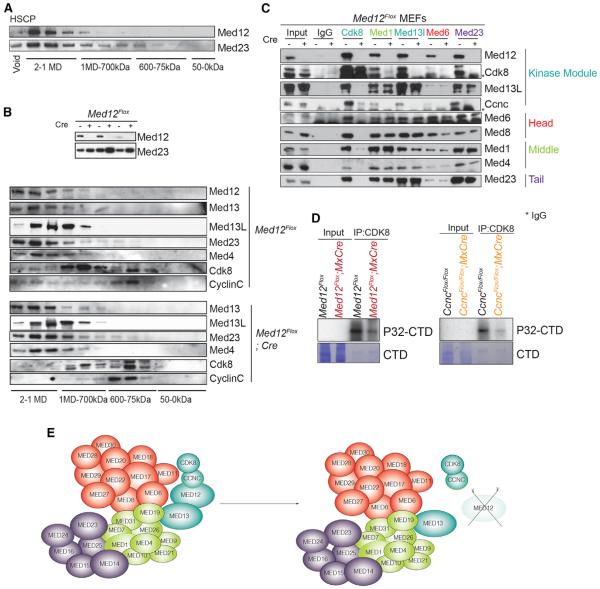
To test if MED12 exists independently from Mediator in HSPCs, we fractionated nuclear extracts of an HSPC-like cell line, HPC-7 (Pinto do O et al., 1998), by gel filtration chromatography, and we observed MED12 associated with Mediator core in large molecular complexes and no free MED12 (Figure 3A). Immunoprecipitation (IP) experiments with different members of the complex confirmed that MED12 and CDK8 bind to the core complex through either MED13 or its paralog MED13L (Figure S3L).
Figure 3. MED12 Loss Leads to Dissociation of CYCLIN C/CDK8 without Affecting the Core Mediator Complex.
(A) Western blot of MED12 and MED23 after Superose 6 size exclusion chromatography on HPC-7 nuclear cell extracts is shown.
(B) Western blot of indicated members of the Mediator complex components after Superose 6 size exclusion chromatography on nuclear extracts of control or MED12-deficient MEFs. Upper part shows MED12 deletion in the first eluted fractions.
(C) IP was performed with indicated Mediator members.
(D) IP kinase experiments with the indicated mice are shown.
(E) Schema depicting lack of CYCLIN C/CDK8 binding to Mediator core after MED12 deletion.
See also Figure S3.
To better understand MED12 function at the molecular level, we used immortalized MED12-deleted and control MEFs from Med12Flox animals. Gel filtration chromatography of nuclear extracts of control MEFs showed that, similar to hematopoietic cells, MED12 co-elutes in high-molecular-weight fractions with the Mediator complex, in agreement with previous studies (Figure 3B) (Knuesel et al., 2009b). A fraction of CDK8 associated with CYCLIN C but did not co-elute with MED12; likewise, MED12 coexists with the Mediator complex without CDK8/CYCLIN C, suggesting independent functions (Knuesel et al., 2009b). Analysis of gel filtration fractions from MED12-depleted MEFs showed that the Mediator core remained unaffected upon MED12 loss but CDK8 and CYCLIN C failed to bind the core complex (Figure 3B).
IP experiments with representative members of each Mediator module showed that, in the absence of MED12, MED13/MED13L remained bound to the core complex while CDK8 and CYCLIN C failed to bind (Figure 3C). Interactions among the head, middle, and tail modules remained unaltered in MED12-deficient cells. In vitro IP kinase experiments with anti-CDK8 revealed that kinase activity was negatively impacted in both MED12-deficient and CYCLIN C-deficient bone marrow cells (Figure 3D). These experiments demonstrated that MED12 functions as part of the Mediator complex and that MED12 deletion leads to detachment of CDK8/CYCLIN C from the core complex and reduction of kinase activity (Figures 3E and S3M). As deletion of CDK8/CYCLIN C did not affect HSC function, these results suggest that MED12 may bring additional function(s) to the Mediator complex necessary for the maintenance of hematopoiesis.
MED12 Deletion Leads to the Loss of HSC Gene Signatures and Affects Cell Survival
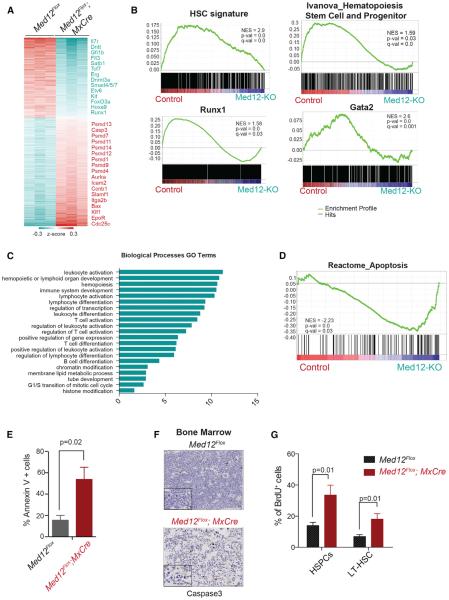
HSPCs from MED12-deficient animals had an aberrant immunophenotypic profile characterized by low surface levels of the essential SCF receptor c-KIT (Figure 1L). To uncover additional molecular signatures related to this phenotype, we performed transcriptome profiling using mRNA isolated from Med12Flox; MxCre HSPCs 4 days after pI:pC injection (Figure 4A). This strategy allowed us to recover MED12-deficient HSPCs before they were eradicated from the bone marrow. We observed that the top downregulated genes upon Med12 deletion included essential transcription required for HSC maintenance (e.g., Erg, Etv6, Runx1, and Tcf7) and essential HSC and progenitor cell surface markers (e.g., c-Kit, Flt3, and Il7r). Among upregulated genes, we found several members of the proteasome pathway (e.g., Psmd1, Psmd12, and Psmd14), regulators of the cell cycle (e.g Cdc25c and Ccnb1), and a large number of regulators of the megakaryocyte-erythroid progenitor lineage (EpoR, Itga2b, and Klf1). Gene set enrichment analysis (GSEA) confirmed that HSC gene signatures and specific gene modules controlled by key hematopoietic transcription factors, such as RUNX1, GATA2, or FLI1/ERG, were significantly under-represented in MED12-deficient HSPCs (Figure 4B; data not shown), suggesting that MED12 plays a role in the function of these putative-associated transcription factors in HSC homeostasis. Gene ontology analysis of downregulated genes (Figure 4C) revealed that many essential biological processes of hematopoiesis were significantly represented, while upregulated genes showed more general and housekeeping cellular functions (Figure S4A).
Figure 4. MED12 Deletion Causes Loss of HSC Signatures and Increased Rates of Apoptosis.
(A) Differentially expressed genes (fold change >1.5, false discovery rate [FDR] p value < 0.05) between Med12Flox;Mx-Cre and Med12Flox HSPCs 4 days after pI:pC are shown.
(B) HSC and key hematopoietic transcription factor gene signatures are decreased upon MED12 deletion, as determined by GSEA.
(C) Biological function annotation of downregulated genes in MED12-deficient HSPCs, using The Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al., 2009), is shown.
(D) Representative apoptosis signature identified by GSEA enriched in MED12-deficient cells is shown.
(E) Annexin V staining in indicated mice 4 days after pI:pC is shown.
(F) Cleaved caspase-3 immunohistochemistry in bone marrow of Med12Flox;Mx-Cre and Med12Flox mice 10 days after pI:pC. Lower panel shows higher magnification.
(G) In vivo BrdU+ labeling of LT-HSCs (Lineage−, c-Kit+, Sca-1+, CD150+, and CD48−) and HSPCs (Lineage, c-Kit+, and Sca-1+) is shown. Scale bars, 100 and 10 μm.
Data represent mean ± SD; p values were determined with two-tailed Student's t tests. See also Figure S4 and Table S5.
Apoptotic gene signatures were positively enriched in MED12-deficient cells (Figure 4D), which was supported by Ingenuity Pathways analysis (Figure S4B). Subsequent Annexin V assays further confirmed that MED12-deficient HSPCs from Med12Flox; MxCre mice had higher apoptotic rates than control HSPCs from littermate animals (Figure 4E). Immunohistochemistry of bone marrow sections of MED12-deficient mice also showed an increase in cells positive for cleaved caspase-3 (Figure 4F). Alteration of cell cycle-related genes (Figure S4C) led us to also investigate cell-cycle changes. In line with the gene expression changes, bromodeoxyuridine (BrdU) assays uncovered an increase in proliferation rates in MED12-deficient HSPCs (Figure 4G), which was confirmed by Ki-67/DAPI assays showing an elevated number of HSPC cells in S-G2/M cells (Figure S4D), most likely as a response to the massive loss of bone marrow cells. To assess if the survival and proliferation phenotypes observed were independent of MED12 binding to CYCLIN C/CDK8, we performed Annexin V and Ki-67/DAPI staining in HSPCs from CYCLIN C-deficient mice, and we corroborated that these animals did not present obvious defects in apoptosis or cell-cycle activity (Figures S4E and S4F).
MED12 Is a Key Element of HSC-Specific Enhancers
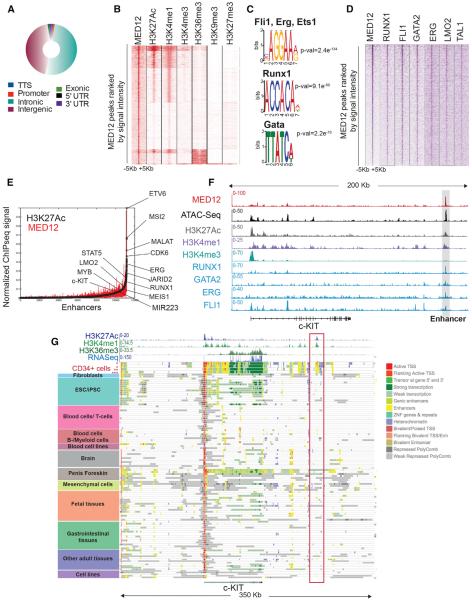
Our results showed that Med12 deletion alters HSC-specific gene expression programs and affects HSPC survival. To understand the molecular mechanisms behind this rapid response, we performed chromatin IP sequencing (ChIP-seq) in primary human CD34+ HSPCs. Genomic distribution analysis of the ~5,000 high-confidence MED12 peaks showed that the majority of the MED12-associated regions were outside promoters (Figure 5A; Table S1), with 84% of loci located distally farther than 10 kb away (Figures S5A and S5B). MED12-bound regions correlated to genes that were both up- and downregulated; however, a slightly higher correlation of MED12 peaks was observed with genes downregulated upon Med12 deletion (Figures S5C and S5D). Genomic Regions Enrichment of Annotations Tool (GREAT) analysis of downregulated genes associated to a MED12-bound region identified mouse phenotypes correlated to hematopoietic and immune cell abnormalities (Figure S5E), while upregulated genes were not associated with any specific murine phenotype. Further comparison with ChIP-seq experiments of diverse histone marks in CD34+ cells available from Human Reference Epigenome Mapping Project (Table S2) revealed that MED12 mostly co-localized with histone modifications characteristic of active enhancers (H3K4me1 and H3K27Ac) (Figure 5B). We detected no MED12 binding within actively transcribed gene bodies (as demonstrated by the absence of H3K36me3) or at repressive polycomb-controlled regions (H3K27me3).
Figure 5. MED12 Colocalizes with Essential Transcription Factors at HSPC-Specific Active Enhancers.
(A) Genomic distribution of MED12-occupied regions in human CD34+ cells is shown.
(B) Heatmap displays MED12 ChIP-seq data and indicated histone marks clustered by k-means, all arranged by MED12 signal density.
(C) Representative transcription factor-binding motifs enriched at MED12-bound regions are shown.
(D) Heatmap shows MED12 ChIP-seq data and transcription factors sorted by MED12 peaks.
(E) Distribution of H3K27Ac and MED12 ChIP-seq density across enhancers and super-enhancers in human CD34+ cells is shown.
(F) Track image shows the indicated ChIP-seq and assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq) experiments on the c-KIT locus.
(G) Gene track image shows ChromHMM states at c-KIT locus in all tissues available from Roadmap Epigenomics project (http://www.roadmapepigenomics.org/). *mobilized CD34+; **cultured CD34+ cells; ***primary CD34+ cells.
Computational analysis of MED12-occupied regions identified DNA sequence motifs of essential hematopoietic transcription factors, such as FLI1, RUNX1, or GATA (Figure 5C). Comparison with previously reported ChIP-seq data on hematopoietic transcription factors in CD34+ cells confirmed that MED12 colocalizes with key transcription factors, such as RUNX1, FLI1, GATA2, or ERG and, to a lesser extent, with LMO2 and TAL1 (Figure 5D; Table S2) (Beck et al., 2013). Interestingly, gene signatures regulated by these transcription factors were negatively affected by Med12 deletion in the mouse hematopoietic system (Figure 4B), suggesting that they may function with MED12 on chromatin to execute their transcriptional program.
Although MED1 has been shown to be localized at enhancers and super-enhancers (Whyte et al., 2013), little is known about the functional role of distinct Mediator members in enhancers of adult stem cells. To investigate if MED12 is part of such structures, we defined enhancers and super-enhancers of human HSPCs by ranking their H3K27Ac signal density (Whyte et al., 2013), and we interrogated MED12 signal density on these regions. Key hematopoietic genes were associated to super-enhancers (ETV6, ERG, KIT, LMO2, and MEIS1), and MED12 was present on those regions (Figure 5E). There was a high degree of overlap between human and mouse MED12-occupied enhancers (Figure S5F). We closely examined several loci of essential HSC genes, such as c-KIT or FLT3, and we observed that MED12-occupied enhancers were located at accessible chromatin regions and co-occuppied with key hematopoietic transcription factors and histone marks indicative of active enhancers (H3K27Ac and H3K4me1) (Figures 5F and S5G). We identified a MED12-bound putative enhancer element 160 kb 3° of c-KIT. Interestingly, available chromatin state data (ChromHMM; Ernst and Kellis, 2012) revealed that this element was exclusively active in human CD34+ cells (Figure 5G), suggesting that MED12 marks cell-type-specific active enhancers. Taken together, these results show that MED12 colocalizes with key hematopoietic transcription factors at active enhancers of HSPCs, and they suggest that MED12 coordinates the activity of cardinal transcription factors at enhancers.
MED12 Loss Results in Enhancer Inactivation
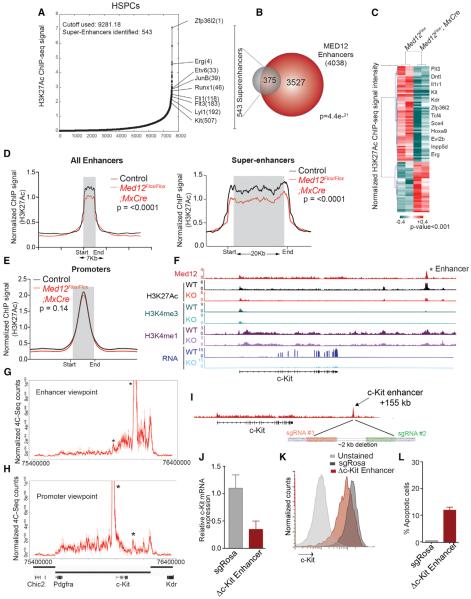
Most work related to Mediator function has focused on transcription initiation and elongation (Allen and Taatjes, 2015; Malik and Roeder, 2010). Only a handful of studies have addressed the Mediator role at enhancers, and these mainly address the contribution of Mediator to chromatin architecture and interaction with non-coding RNAs (Kagey et al., 2010; Lai et al., 2013). Many studies have been performed in in vitro systems, thus hindering the understanding of Mediator function at enhancers in vivo. Since we found MED12 primarily located at enhancers of human HSPCs, we sought to investigate the mechanistic consequences of MED12 depletion at HSPCs in vivo. Due to the rare number of HSPCs per mouse, we used an indexing-first chromatin IP approach coupled with next-generation sequencing (iChIP-seq) to map the H3K27Ac, H3K4me1, and H3K4me3 histone marks from 20,000 HSPCs from MED12-deficient and control littermates (Lara-Astiaso et al., 2014). H3K27Ac revealed 7,560 enhancers in wild-type HSPCs, from which 543 were classified as super-enhancers and were mostly associated with key regulators of HSPC function (Figure 6A; Table S3). Super-enhancer-associated genes were involved in cellular functions required for optimal hematopoiesis (Figures S6A and S6B).
Figure 6. MED12 Is Required to Maintain Enhancer Activity In Vivo.
(A) H3K27Ac ChIP-seq density across enhancers and super-enhancers in mouse HSPCs is shown.
(B) Overlap between super-enhancers in mouse-isolated HSPCs and MED12-bound enhancers identified in HPC-7. MED12 ChIP-seq was performed in HPC-7 cells due to the requirement of a large number of cells. Some super-enhancers contain more than one MED12 enhancer.
(C) Super-enhancer regions showing significant changes in H3K27Ac density after Med12 deletion. Some relevant hematopoietic genes with an associated super-enhancer are indicated. Average of experiments of iChIP HSPCs from four independent Med12Flox and Med12Flox;Mx-Cre mice is shown. Analysis was done with DiffBind, which calculates the p value with the Wald test.
(D) ChIP-seq density profile representing H3K27Ac signal at enhancer and super-enhancers is shown.
(E) ChIP-seq density profile showing H3K27Ac signal at promoters. The p values were calculated with the Mann-Whitney U test for (D) and (E).
(F) Track image of the indicated ChIP-seq binding profiles at the c-Kit locus Med12Flox;Mx-Cre and Med12Flox mice 4 days after pI:pC injection. MED12 ChIP-seq was done on HPC-7 and histone marks were profiled in mouse-derived HSPCs.
(G and H) High-resolution 4C-seq performed with c-Kit promoter and enhancer baits in HPC-7 cells. Black bars indicate TADs (Dixon et al., 2012). Full lines represent the mean of replicates and dashed lines represent the signal for each replicate. Asterisks show the tested areas.
(I) Schematics show c-Kit enhancer region deleted.
(J) c-Kit mRNA levels upon enhancer deletion are shown.
(K) c-KIT surface levels marker upon enhancer deletion is shown.
(L) Annexin V staining upon c-KIT enhancer deletion.
Error bars represent mean ± SD; p values were determined with two-tailed Student's t tests. See also Figure S6 and Table S3.
An unbiased comparison with MED12-associated regions confirmed that a highly significant number (almost 70%) of H3K27Ac-defined super-enhancers bear MED12 (Figures 6B and S6C). Super-enhancer-associated genes suffered a higher expression reduction than enhancer-associated genes upon MED12 deletion in vivo (Figure S6D). Interestingly, shortly after pI:pC injection, a large number of the super-enhancer regions showed reduced H3K27Ac density, suggesting that their activation state was diminished (Figure 6C). There was a high degree of overlap (86 loci, p [x≥86] = 5.9e–36) between HSPCs with differential H3K27Ac load and MED12-occupied regions. Enhancer ranking by H3K27Ac density in HSPCs further revealed a reduction in H3K27Ac signal at enhancers of MED12-deficient HSPCs, which was more pronounced at super-enhancers (Figures 6D and S6E). H3K27Ac levels were not significantly decreased at promoter regions (Figure 6E), supporting a role for MED12 in the maintenance of active enhancers of key hematopoietic genes. Global levels of H3K27Ac were reduced in mouse-isolated HSPCs and the HPC-7 cell line upon MED12 deletion (Figure S6F). Activity of transcription start sites (TSSs) was not globally affected; however, the levels of H3K27Ac and H3K4me3 at TSSs of super-enhancer-associated genes were decreased, albeit to a lower degree than super-enhancers (Figure S6G). These results indicate an enhancer-specific role for MED12 in preserving H3K27Ac levels for adequate enhancer activity.
To better understand MED12 function in HSPCs, we focused on the c-Kit gene since its expression was rapidly reduced upon MED12 depletion (Figures 1K and 1L). We mapped in mouse HSPCs the distant c-KIT enhancer element that we identified in human CD34+ cells, and we confirmed that it carried both H3K27Ac and H3K4me1 marks and was equally occupied by MED12. Med12 deletion in vivo led to a significant loss of H3K27Ac from this enhancer (Figure 6F), similar to several other essential hematopoietic genes, such as Flt3 (Figure S6H), Hoxa9, or Erg (data not shown). On the other hand, non-hematopoietic-specific (housekeeping) genes remained mainly unaffected (Figure S6I). To demonstrate that this region was a c-Kit enhancer, we used high-resolution circular chromosomal conformation capture sequencing (4C-seq), and we interrogated chromatin interactions inside the c-Kit-containing topological association domain (TAD) in HPC-7 cells. We used two 4C-seq viewpoints, c-Kit TSS and the conserved mouse +155 kb MED12-bound enhancer. In both, DNA interactions were largely confined within the TAD and decreased outside the TAD boundaries. Promoter-enhancer interactions could be detected using either viewpoint (Figures 6G and 6H). Upstream interactions from the enhancer were sharply delimited by the c-Kit gene (Figure 6G). To demonstrate that this enhancer element controls c-Kit expression, we used CRISPR-mediated deletion in a leukemia cell line (RN2) with stem cell characteristics, including surface expression and dependence on c-KIT signaling for growth (Somervaille and Cleary, 2006) (Figure 6I). RN2 cells also have the same active +155 kb active c-Kit enhancer based on our H3K27Ac ChIP-seq analysis (data not shown), and stable Cas9-expressing RN2 cells were readily available. A 2-kb deletion flanking the MED12 peak confirmed that this region contains a bona fide enhancer required to maintain c-Kit expression at the mRNA (Figure 6J) and, consequently, at the protein level (Figure 6K). c-Kit downregulation caused by CRISPR-mediated deletion of this enhancer also led to increased levels of apoptosis (Figure 6L).
MED12 Interacts and Cooperates with P300 at the Chromatin
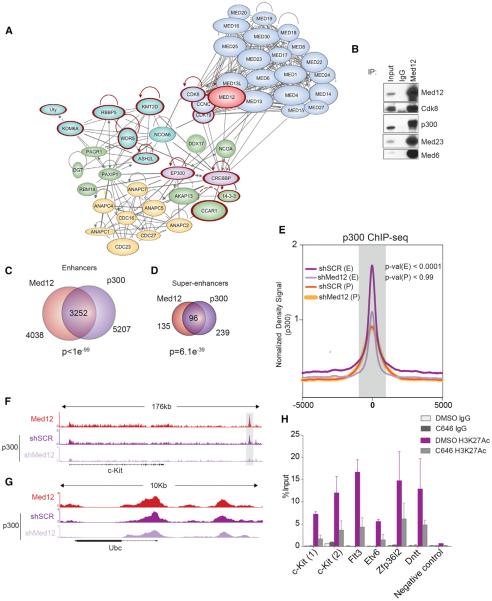
To identify mechanisms by which MED12 preserves hematopoietic enhancer activity, we performed IP experiments coupled to mass spectrometry using HPC-7 cells (Figure S7A). As expected, MED12 associated with most members of the Mediator complex. MED12 complexes were devoid of MED26, as previously reported (Paoletti et al., 2006), and of MED12L, which is mutually exclusive with MED12 (Daniels et al., 2013) (Figure 7A; Table S4). MED13 and MED13L were among the highest represented proteins, confirming that MED12 can interact with Mediator through both of them. Strikingly, we found a high abundance of the two main histone acetylases targeting H3K27 (P300 and CBP) (Sun et al., 2015), suggesting that they could collaborate with MED12 in regulating H3K27Ac levels at enhancers. Interestingly, we also found several members of the ASCOM complex (KMT2D, RBBP5, WDR5, ASHL2, NCOA6, and KDM6A) (Goo et al., 2003), which combines activating histone methyltransferase activity on lysine 4 and repressing histone demethylase activity on lysine 27. Other interaction partners included several RNA-binding proteins, DNA/protein modifiers, and members of the anaphase-promoting complex (APC).
Figure 7. MED12 Interacts with P300 and Promotes Its Binding to the Chromatin on Hematopoietic Enhancers.
(A) MED12 interaction partners network in HPC-7 cells. Proteins with mutations in hematological diseases are highlighted in red.
(B) MED12 IP in HPC-7 confirming interaction with P300. A repetition of this experiment is shown in Figure S7B.
(C) Overlap of MED12-associated and P300-associated enhancer regions is shown.
(D) Overlap of MED12-associated and P300-associated super-enhancer regions is shown.
(E) Normalized P300 ChIP-seq signal profiles of P300-MED12 overlapping enhancers centered on distal P300 and promoters. The p values were calculated with the Mann-Whitney U test.
(F) ChIP-seq track of p300 and MED12 on c-Kit enhancer locus is shown.
(G) ChIP-seq track of P300 and MED12 on Ubc locus is shown.
(H) ChIP-qPCR analysis of H3K27Ac at the indicated enhancers in mouse-isolated HSPCs following a 2-hr treatment with DMSO or with 10 μM C646 nM.
Error bars represent mean ± SD. See also Figure S7, Table S1, and Table S4.
The significant changes on H3K27Ac load at key HSPC gene enhancers and the proteomics results prompted us to investigate if MED12 cooperates with P300 to maintain enhancers in an active state. Upon confirming that endogenous MED12 and P300 robustly interact in HPC-7 cells (Figures 7B and S7B), we performed an unbiased analysis of putative enhancer regions by independently identifying enhancers based on MED12 or P300 ChIP-seq levels. We observed that MED12 and P300 overlap in 80% of enhancers and >70% of super-enhancers (Figures 7C and 7D). Gene ontology analysis of overlapping loci confirmed that these regions are relevant to hematopoietic function (Figures S7C and S7D). DNA sequence motif search of MED12 and P300 overlapping regions revealed specific motifs of key hematopoietic genes (Figure S7E). Most importantly, P300 ChIP-seq experiments upon MED12 depletion showed that P300 was depleted from enhancers in the absence of MED12 (Figure 7E), without causing a total P300 reduction (Figures S7F and S7G). Interestingly, P300 levels at the c-Kit enhancer were decreased upon MED12 silencing (Figure 7F), but P300 loss was not noticed in many non-hematopoietic genes like Ubc (Figure 7G). These results indicated that P300 loss upon MED12 deletion may be the cause of enhancer inactivation in MED12-deficient HSPCs. Supporting this notion, the addition of a P300-specific inhibitor (C646) led to an H3K27Ac reduction at loci regulated by MED12 (Figure 7H). These results demonstrate that MED12 interacts with P300 and promotes its stability at the chromatin and the deposition of H3K27ac. These results provide a mechanistic insight for the maintenance of lineage-specific enhancers in key HSC genes.
DISCUSSION
We identify an essential role for MED12 as a regulator of HSC homeostasis in vivo. Using different Cre strains and in vitro systems, we demonstrate that MED12 deletion leads to a rapid loss of HSPCs, causing acute bone marrow aplasia and lethality. MED12 deletion negatively affects key HSC gene expression signatures, and, consequently, it induces an increased rate of apoptosis in HSPCs. MED12 and the CYCLIN C/CDK8 kinase module have been connected to both activating and repressing functions on gene transcription, depending on cellular context (Akoulitchev et al., 2000; Allen and Taatjes, 2015; Carrera et al., 2008; Galbraith et al., 2013). While we do not claim that in this system MED12 has exclusively activating functions, we focused on MED12 activating functions, as our ChIP-seq analysis revealed colocalization with histone marks, transcription factors, and P300, all characteristic of active enhancers. Also, we did not observe any correlation of MED12 binding with available repressive histone marks (H3K27me3 and H3K9me3). Moreover, we did not notice a strong binding of MED12 at line-age-specific genes that were upregulated upon MED12 deletion (data not shown). Nevertheless, it would be interesting to address repressive functions of MED12 in such lineage-specific stem cell systems.
We show that MED12 co-localizes with key hematopoietic transcription factors, such as RUNX1, GATA2, or FLI1, that are likely to recruit MED12 to the chromatin, where it interacts and cooperates with additional transcriptional co-activators to maintain enhancers of essential hematopoietic genes active. Supporting this notion, we show the cooperation of MED12 with P300 at enhancers, including at the c-Kit enhancer. Thus, we believe that MED12 provides an essential platform among co-activators, transcription factors, and the Mediator complex required to maintain the active state of hematopoietic-specific enhancers. Similar to previous reports, we support the idea that MED12 function depends on the biological context and lineage specificity could be provided by distinct interactions with lineage-specific transcription factors and co-activators (Allen and Taatjes, 2015). We thus predict that MED12 loss would lead to distinct outcomes, depending on cell type and differentiation stage. In agreement, MED12 deletion in MEFs or pluripotent ESCs has no discernible phenotypes. Most importantly, our preliminary studies have shown that Med12 deletion in hair follicle stem cells does induce cell death (as in HSCs) but causes a reduction of cell proliferation in epidermal progenitor cells, as indicated by the loss of Ki-67 staining. This is an example of how two different types of stem and progenitor cells sense MED12 deletion differently: by initiating cell death (HSC) or by suppressing cell-cycle entry (hair follicle). It will be fascinating to know in the future which (and how) transcription factors directly cooperate with MED12 to control diverse biological functions in distinct stem cell systems.
Unlike deletion of MED12, loss of MED1 in vivo does not affect HSC homeostasis (Stumpf et al., 2010). Interestingly, we found that deletion of other members of the kinase module (MED13, CYCLIN C, and CDK8) did not affect the survival of HSPCs, suggesting that MED12 is indispensable for the maintenance of hematopoiesis. This could be explained in part by redundancy between members of the complex. For instance, both MED13 and MED13L physically connect the kinase module with the Mediator core (Knuesel et al., 2009a). Moreover, while CDK8 and CDK19 have non-redundant functions (Galbraith et al., 2013), it is necessary to target both to affect cell survival of myeloid leukemia cells (Pelish et al., 2015). However, the mere existence of paralogs cannot account for the MED12 functions herein described. Indeed, there are no known paralogs for CYCLIN C. These data and our recently published work show that CYCLIN C deletion not only has no effect on HSPC survival, but also leads to the induction of acute leukemia, supporting the notion that CYCLIN C can function as a tumor suppressor (Li et al., 2014). All these are in agreement with our findings showing kinase module-independent functions, like P300 binding, that could explain the role of MED12 in hematopoiesis. We should mention that the MED12 paralog MED12L is almost exclusively expressed in LT-HSC, MPP1, and megakaryocyte progenitors (unpublished data; http://www.immgen.org). Since MED12 and MED12L are mutually exclusive at binding the Mediator core complex (Galbraith et al., 2013), it would be intriguing to test whether these two subunits coordinately regulate early stages of HSC and progenitor differentiation by regulating distinct transcriptional programs.
Interaction of the kinase module with the Mediator core is partly regulated post-translationally. Recently, we and others have shown that post-translational modification of MED13/MED13L and MED12 by the ubiquitin ligase FBXW7 controls protein stability and the binding of the kinase module with the core Mediator (Davis et al., 2013). FBXW7 previously has been shown to be an essential HSPC regulator (Matsuoka et al., 2008; Thompson et al., 2008). Since FBXW7 controls the half-life or key hematopoietic transcription factors like NOTCH1/2, c-MYC, or CEBPA (Welcker and Clurman, 2008), it tempting to ask about the possibility that FBXW7 loss/mutations could affect both specific transcription factors and their associated Mediator complexes on key hematopoietic enhancers.
A number of studies have identified MED12 somatic mutations in blood cancers, such as chronic lymphocytic leukemia (CLL) or acute myeloid leukemia (AML) (Kämpjärvi et al., 2015; Landau et al., 2013). Recent studies of mutations targeting the N-terminal end of MED12 in uterine leyomyomas suggested that such MED12 mutations could be gain of function (Mittal et al., 2015). Since mutations driving CLL also reside in the N terminus (Kämpjärvi et al., 2015), it is tempting to speculate that MED12 gain-of-function mutations could give rise to leukemia, as MED12 loss of function would lead to bone marrow failure according to our research. Interestingly, several of the co-activators interacting with MED12 in HSPCs are also frequently mutated in leukemia and lymphoma, including P300, CBP, KMT2D, WDR5, and KMD6A (Figure 7A, red circle) (Giotopoulos et al., 2016; Ntziachristos et al., 2012; Ortega-Molina et al., 2015). Future work will test whether these genetic events induce cellular transformation by modulating enhancer-promoter interactions and transcription factor activity.
EXPERIMENTAL PROCEDURES
Additional information and details of procedures used in this study can be found in the Supplemental Experimental Procedures.
Methylcellulose Assays
Bone marrow Med12Flox;MxCre or control mice were plated on cytokine-supplemented methylcellulose medium, and the number and morphology of colonies were scored 7 days later. Alternatively, for HSPC assays, bone marrow cell Lin−, Sca1, and c-kit+ were sorted with a fluorescence-activated cell sorting (FACS) aria. Sorted cells were infected with retrovirus expressing pMig-IRES-GFP or pMIG-Cre-GFP or a PLKO-puro-IRES-GFP with specific shRNAs for Cdk8, Med12, or a scrambled control. Then 24 hr post-infection, GFP+ cells were sorted and 500 or 1,000 cells were plated in methylcellulose. Colony numbers were assessed in 7–10 days. Animal experiments were performed in accordance with protocols approved by the New York University Langone Medical Center Institutional Animal Care and Use Committee.
IP Kinase Assay
Nuclear extracts of bone marrow cells from Med12Flox, Med12Flox;MxCre, CcncFlox/Flox, and CcncFlox/Flox;MxCre were used for anti-CDK8 IPs, and, sub-sequently, kinase assays were performed with 150 ng GST-CTD in a final volume of 20 μl kinase buffer containing 0.02 mCi [γ-32P] ATP for 1 hr.
Microarray and GSEA
Microarrays were performed with RNA isolated from HSPCs of individual mice, and theMOE430 Plus 2 oligonucleotide arrays were used.
iChIP
iCHIP was performed as previously described (Lara-Astiaso et al., 2014). Sonicated chromatin from control and MED12-deficient mouse HSPCs was bar-coded on beads after IP with anti-H3 antibody. Samples were pooled and further subjected to IP with antibodies against specific histone marks. Immunoprecipitated DNA was isolated and amplified with Illumina-compatible primers. Libraries were sequenced in a HiSeq2500 instrument.
Statistical Analysis
The p values were calculated using unpaired Student's t test expressed as mean ± SEM or mean ± SD as indicated. GraphPad Prism software was used unless otherwise indicated. Hypergeometric test was used to calculate p values in Venn diagrams. Other p values for specific figures were calculated with the indicated programs used.
Supplementary Material
Highlights.
MED12 is required for hematopoietic stem cell function in a cell-autonomous manner
Depleting other members of the Mediator kinase module does not affect HSC survival
MED12 deletion leads to H3K27Ac loss at enhancers of key HSC genes, such as c-Kit
MED12 cooperates with P300 at enhancers of essential hematopoietic genes
ACKNOWLEDGMENTS
We thank all members of the I.A. lab for useful discussions; A. Heguy and the NYU GTC (NIH grant P30CA016087-30) for expertise with sequencing experiments and library preparation; Dr. B. Ueberheide at the NYU Proteomics Resource Center (Cancer Center Support Grant P30CA016087) for mass spectrometry experiments; Igor Dolgarev, Harris Lazaris, and Yixiao Gong for helpful bioinformatic discussions and protocols; and The High Performance Computing Facility of the NYU Center of Health Informatics and Bioinformatics. We thank the Immunohistochemistry and Histopathology and Immunohistochemistry cores and the Cytometry and Cell Sorting facility at NYU (both supported by the Cancer Center Support Grant P30CA016087). We thank E.N. Olson and R. Bassel-Duby at the University of Texas Southwestern Medical Center for kindly providing the Med13Flox/Flox mice. We thank Dr. S. Heim feld in Fred Hutchinson Cancer Research Center for providing human CD34+ cells (Core Center of Excellence NIDDK Grant DK56465), Dr. Chris Vakoc for the RN2-Cas9 line, and Professor L. Carlsson (University of Umeå) for the HPC-7 cells. The I.A. lab is supported by the NIH (RO1CA133379, RO1CA105129, RO1CA149655, 5RO1CA173636, 1RO1CA194923, and R01 CA190509 to P.S.), the Leukemia and Lymphoma Society (LLS TRP Program), and the NYSTEM program of the New York State Health Department (NYSTEM-N11G-255). B.A.-O. was supported by the Feodor Lynen Fellowship from the Humboldt-Foundation and the Deutsche José Carreras LeukÄmie-Stiftung e.V.
Footnotes
ACCESSION NUMBERS The accession number for the data reported in this paper is GEO: GSE76055.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2016.08.004.
AUTHOR CONTRIBUTIONS B.A.-O. designed, performed, and analyzed most experiments. R.S.-M. and B.A.-O. performed gel filtration chromatography, mouse ChIP-seq, and IP kinase. E.W. performed CRISPR enhancer deletion. E.T. performed ChIP-seq in human CD34+ cells. A.C. provided assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) experiments and L.I.Z. provided human CD34+ expertise. S.L., J.M., N.Z., M.S.-D., K.W., C.K., and M.G. provided technical assistance. H.S. provided Med12Flox/Flox mice, A.F. and P.S. provided the CcnFlox/Flox mice, and L.A. provided the Med13Flox/Flox mice. P.P.R., R.R., and J.A.S. assisted in 4C-seq experiments. M.S. performed skin stem cell experiments and provided expertise. D.R. provided resources. B.A.-O. analyzed genome-wide data and A.T. assisted in the bioinformatic analysis. B.A.-O. and I.A. conceived ideas and wrote the manuscript.
REFERENCES
- Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D, Thoms JA, Perera D, Schütte J, Unnikrishnan A, Knezevic K, Kinston SJ, Wilson NK, O'Brien TA, Göttgens B, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–e22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. USA. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Ford M, Schwinn MK, Benink H, Galbraith MD, Amunugama R, Jones R, Allen D, Okazaki N, Yamakawa H, et al. Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK-Mediator kinase module. J. Proteom. Bioinform. 2013;S2:004. [Google Scholar]
- Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 2013;27:151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domen J, Weissman IL. Hematopoietic stem cells need two signals to prevent apoptosis; BCL-2 can provide one of these, Kitl/c-Kit signaling the other. J. Exp. Med. 2000;192:1707–1718. doi: 10.1084/jem.192.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol. Cell. 2015;60:328–337. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giotopoulos G, Chan WI, Horton SJ, Ruau D, Gallipoli P, Fowler A, Crawley C, Papaemmanuil E, Campbell PJ, Göttgens B, et al. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene. 2016;35:279–289. doi: 10.1038/onc.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics. 2009;Chapter 13:13.11. doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KÄmpjÄrvi K, JÄrvinen TM, Heikkinen T, Ruppert AS, Senter L, Hoag KW, Dufva O, Kontro M, Rassenti L, Hertlein E, et al. Somatic MED12 mutations are associated with poor prognosis markers in chronic lymphocytic leukemia. Oncotarget. 2015;6:1884–1888. doi: 10.18632/oncotarget.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KÄmpjÄrvi K, Kim NH, Keskitalo S, Clark AD, von Nandelstadh P, Turunen M, Heikkinen T, Park MJ, MÄkinen N, Kivinummi K, et al. Somatic MED12 mutations in prostate cancer and uterine leiomyomas promote tumorigenesis through distinct mechanisms. Prostate. 2016;76:22–31. doi: 10.1002/pros.23092. [DOI] [PubMed] [Google Scholar]
- Keightley MC, Layton JE, Hayman JW, Heath JK, Lieschke GJ. Mediator subunit 12 is required for neutrophil development in zebrafish. PLoS ONE. 2011;6:e23845. doi: 10.1371/journal.pone.0023845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009a;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 2009b;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Fassl A, Chick J, Inuzuka H, Li X, Mansour MR, Liu L, Wang H, King B, Shaik S, et al. Cyclin C is a haploinsufficient tumour suppressor. Nat. Cell Biol. 2014;16:1080–1091. doi: 10.1038/ncb3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22:986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal P, Shin YH, Yatsenko SA, Castro CA, Surti U, Rajkovic A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Invest. 2015;125:3280–3284. doi: 10.1172/JCI81534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, Jiang M, Hu D, Agirre X, Niesvizky I, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 2015;21:1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. USA. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A, Fadeyi O, Christie AL, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto do O P, Kolterud A, Carlsson L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 1998;17:5744–5756. doi: 10.1093/emboj/17.19.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137:2723–2731. doi: 10.1242/dev.053660. [DOI] [PubMed] [Google Scholar]
- Rossi L, Lin KK, Boles NC, Yang L, King KY, Jeong M, Mayle A, Goodell MA. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- Stumpf M, Yue X, Schmitz S, Luche H, Reddy JK, Borggrefe T. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc. Natl. Acad. Sci. USA. 2010;107:21541–21546. doi: 10.1073/pnas.1005794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Man N, Tan Y, Nimer SD, Wang L. The role of histone acetyltransferases in normal and malignant hematopoiesis. Front. Oncol. 2015;5:108. doi: 10.3389/fonc.2015.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell. 2014;157:1430–1444. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen M, Spaeth JM, Keskitalo S, Park MJ, Kivioja T, Clark AD, Mäkinen N, Gao F, Palin K, Nurkkala H, et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014;7:654–660. doi: 10.1016/j.celrep.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.